XSB0944 : integrin beta 3 [Rattus norvegicus]
[ CaMP Format ]
This entry is computationally expanded from SB0043
* Basic Information
| Organism | Rattus norvegicus (Norway rat) |
| Protein Names | Integrin beta; integrin beta 3; integrin beta 3 (Cd61) |
| Gene Names | Itgb3; itgb3; integrin beta 3 |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 787 aa
Average Mass: 86.930 kDa
Monoisotopic Mass: 86.873 kDa
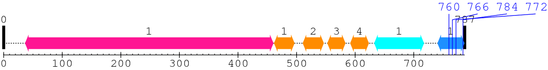
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Integrin_beta 1. | 37 | 460 | 989.5 | 1.4e-294 |
| EGF_2 1. | 462 | 496 | 19.2 | 0.017 |
| EGF_2 2. | 511 | 546 | 31.9 | 2.6e-06 |
| EGF_2 3. | 553 | 583 | 26.5 | 0.00011 |
| EGF_2 4. | 592 | 623 | 23.8 | 0.00071 |
| Integrin_B_tail 1. | 633 | 717 | 125.2 | 2.1e-34 |
| Integrin_b_cyt 1. | 741 | 786 | 106.8 | 7.1e-29 |
| --- cleavage 760 (inside Integrin_b_cyt 741..786) --- | ||||
| --- cleavage 766 (inside Integrin_b_cyt 741..786) --- | ||||
| --- cleavage 784 (inside Integrin_b_cyt 741..786) --- | ||||
| --- cleavage 772 (inside Integrin_b_cyt 741..786) --- | ||||
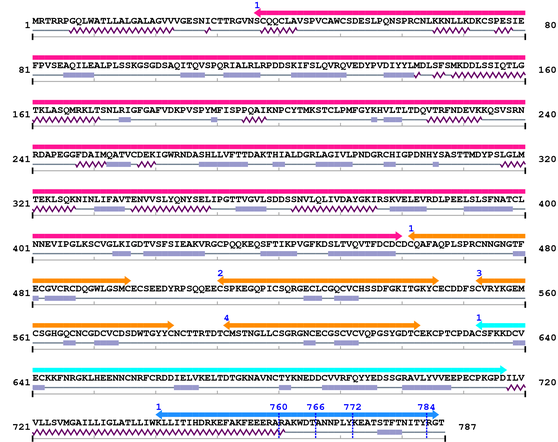
3. Sequence Information
Fasta Sequence: XSB0944.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
4 [sites]
Cleavage sites (±10aa)
[Site 1] EFAKFEEERA760-RAKWDTANNP
Ala760  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu751 | Phe752 | Ala753 | Lys754 | Phe755 | Glu756 | Glu757 | Glu758 | Arg759 | Ala760 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg761 | Ala762 | Lys763 | Trp764 | Asp765 | Thr766 | Ala767 | Asn768 | Asn769 | Pro770 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LIGLATLLIWKLLITIHDRKEFAKFEEERARAKWDTANNPLYKEATSTFTNITYRGT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Mus musculus | 84.70 | 16 | unnamed protein product |
| 2 | Bos taurus | 84.70 | 12 | PREDICTED: similar to platelet glycoprotein IIIa i |
| 3 | Macaca mulatta | 84.70 | 12 | PREDICTED: similar to integrin beta chain, beta 3 |
| 4 | Canis familiaris | 84.70 | 10 | platelet glycoprotein IIIa |
| 5 | Rattus norvegicus | 84.70 | 10 | integrin beta 3 |
| 6 | Sus scrofa | 84.70 | 6 | glycoprotein GPIIIa |
| 7 | synthetic construct | 84.70 | 5 | AF239959_1 hybrid integrin beta 3 subunit precurso |
| 8 | rats, Peptide Partial, 723 aa | 84.70 | 1 | beta 3 integrin, GPIIIA |
| 9 | Canis lupus familiaris | 84.70 | 1 | integrin beta chain, beta 3 |
| 10 | Equus caballus | 84.70 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 11 | N/A | 82.80 | 29 | - |
| 12 | Homo sapiens | 82.80 | 24 | integrin beta chain, beta 3 precursor |
| 13 | Pan troglodytes | 82.80 | 11 | PREDICTED: integrin beta chain, beta 3 isoform 4 |
| 14 | Oryctolagus cuniculus | 80.90 | 2 | glycoprotein IIIa |
| 15 | Monodelphis domestica | 80.50 | 5 | PREDICTED: similar to platelet glycoprotein IIIa |
| 16 | Gallus gallus | 79.30 | 7 | integrin, beta 3 precursor |
| 17 | mice, Peptide Partial, 680 aa | 75.90 | 1 | beta 3 integrin, GPIIIA |
| 18 | Xenopus laevis | 74.70 | 6 | integrin beta-3 subunit |
| 19 | Danio rerio | 70.90 | 13 | hypothetical protein LOC100001836 |
| 20 | Tetraodon nigroviridis | 70.90 | 5 | unnamed protein product |
| 21 | Aedes aegypti | 63.90 | 1 | integrin beta subunit |
| 22 | Drosophila melanogaster | 62.00 | 1 | myospheroid CG1560-PA |
| 23 | Tribolium castaneum | 61.60 | 2 | PREDICTED: similar to CG1560-PA |
| 24 | Ostrinia furnacalis | 61.60 | 1 | integrin beta 1 precursor |
| 25 | Anopheles gambiae | 60.50 | 1 | integrin beta subunit |
| 26 | Anopheles gambiae str. PEST | 60.50 | 1 | ENSANGP00000021373 |
| 27 | Pseudoplusia includens | 58.50 | 1 | integrin beta 1 |
| 28 | Ovis aries | 56.60 | 2 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 29 | Pongo pygmaeus | 55.80 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 30 | Felis catus | 55.80 | 1 | integrin beta 1 |
| 31 | Biomphalaria glabrata | 55.80 | 1 | beta integrin subunit |
| 32 | Xenopus tropicalis | 55.80 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 33 | Crassostrea gigas | 55.10 | 1 | integrin beta cgh |
| 34 | Podocoryne carnea | 54.70 | 1 | AF308652_1 integrin beta chain |
| 35 | Caenorhabditis briggsae | 53.90 | 1 | Hypothetical protein CBG03601 |
| 36 | Caenorhabditis elegans | 53.90 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 37 | Ictalurus punctatus | 52.40 | 1 | beta-1 integrin |
| 38 | Strongylocentrotus purpuratus | 50.80 | 4 | integrin beta-C subunit |
| 39 | Lytechinus variegatus | 50.80 | 1 | beta-C integrin subunit |
| 40 | Pacifastacus leniusculus | 46.20 | 1 | integrin |
| 41 | Apis mellifera | 44.70 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 42 | Manduca sexta | 43.10 | 1 | plasmatocyte-specific integrin beta 1 |
| 43 | Spodoptera frugiperda | 42.70 | 1 | integrin beta 1 |
| 44 | Halocynthia roretzi | 38.90 | 1 | integrin beta Hr2 precursor |
| 45 | Mus sp. | 38.10 | 1 | beta 7 integrin |
| 46 | Schistosoma japonicum | 37.00 | 1 | SJCHGC06221 protein |
| 47 | Oncorhynchus mykiss | 36.60 | 1 | CD18 protein |
| 48 | Acropora millepora | 35.80 | 1 | integrin subunit betaCn1 |
| 49 | Bubalus bubalis | 33.50 | 1 | integrin beta-2 |
| 50 | Capra hircus | 33.50 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 51 | Sigmodon hispidus | 33.10 | 1 | AF445415_1 integrin beta-2 precursor |
| 52 | Ovis canadensis | 32.70 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
Top-ranked sequences
| organism | matching |
|---|---|
| Mus musculus | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 731 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 787 |
| Bos taurus | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| Macaca mulatta | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 732 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| Canis familiaris | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| Rattus norvegicus | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 731 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 787 |
| Sus scrofa | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| synthetic construct | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 732 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| rats, Peptide Partial, 723 aa | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 667 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 723 |
| Canis lupus familiaris | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| Equus caballus | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 |||||||||||||||||| # |||||||||||||||||||||||| Sbjct 728 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 784 |
| N/A | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 ||||| |||||||||||| # |||||||||||||||||||||||| Sbjct 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| Homo sapiens | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 ||||| |||||||||||| # |||||||||||||||||||||||| Sbjct 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| Pan troglodytes | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 ||||| |||||||||||| # |||||||||||||||||||||||| Sbjct 732 LIGLAALLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| Oryctolagus cuniculus | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 ||| | |||||||||||| # |||||||||||||||||||||||| Sbjct 732 LIGFALLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYRGT 788 |
| Monodelphis domestica | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYR 785 |||||||||||||||||| # |||||||||||||||||||||| Sbjct 707 LIGLATLLIWKLLITIHDRREFAKFEEERA#RAKWDTANNPLYKEATSTFTNITYR 761 |
| Gallus gallus | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 ||||| |||||||||||| # ||| ||||||||||||||||||| Sbjct 724 LIGLAALLIWKLLITIHDRREFARFEEEKA#RAKWDTGNNPLYKEATSTFTNITYRG 779 |
| mice, Peptide Partial, 680 aa | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNIT 783 |||||||||||||||||| # |||||||||||||||||||| Sbjct 628 LIGLATLLIWKLLITIHDRKEFAKFEEERA#RAKWDTANNPLYKEATSTFTNIT 680 |
| Xenopus laevis | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 |||| |||||||||||| # ||||+||||| |||||||||||| Sbjct 730 LIGLVALLIWKLLITIHDRREFAKFEEERA#KAKWDTAHNPLYKGATSTFTNITYRG 785 |
| Danio rerio | Query 732 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 +||| |||||||||||| # |+| +||||| |||||||||||| Sbjct 734 LGLAALLIWKLLITIHDRREFAKFEEERA#RAKWETGHNPLYKGATSTFTNITYRG 788 |
| Tetraodon nigroviridis | Query 732 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 +||| |||||||||||| # ||| +||||| |||||||||+|| Sbjct 748 LGLAALLIWKLLITIHDRREFAKFEEERA#RAKWDTGHNPLYKGATSTFTNITFRG 802 |
| Aedes aegypti | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 ||||| ||+|||| |||| # ||| ||+||+||+|| | || | Sbjct 779 LIGLAVLLLWKLLTTIHDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTYAG 834 |
| Drosophila melanogaster | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 |+||| ||+|||| |||| # ||| ||+||+||||| | | | Sbjct 790 LVGLAILLLWKLLTTIHDRREFARFEKERM#NAKWDTGENPIYKQATSTFKNPMYAG 845 |
| Tribolium castaneum | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 |+|+| ||+||| |||| # ||| ||+||+||||| | || | Sbjct 773 LLGMAILLLWKLFTTIHDRREFAKFEKEAM#MARWDTGENPIYKQATSTFKNPTYAG 828 |
| Ostrinia furnacalis | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 |+||| |++||++ |||| # ||| ||+||+||||| | || | Sbjct 775 LVGLALLMLWKMVTTIHDRREFARFEKERM#MAKWDTGENPIYKQATSTFKNPTYAG 830 |
| Anopheles gambiae | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 |||+| ||+||+| +||| # ||| ||+||+||+|| | || | Sbjct 781 LIGMAVLLLWKVLTSIHDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTYAG 836 |
| Anopheles gambiae str. PEST | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 |||+| ||+||+| +||| # ||| ||+||+||+|| | || | Sbjct 781 LIGMAVLLLWKVLTSIHDRREFARFEKERM#MAKWDTGENPIYKQATTTFKNPTYAG 836 |
| Pseudoplusia includens | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 |+||| |++||+ | || # ||| ||+||+||||| | || | Sbjct 781 LVGLALLMLWKMATTSHDRREFARFEKERM#MAKWDTGENPIYKQATSTFKNPTYAG 836 |
| Ovis aries | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYR 785 |||+| | |||||++ || # | | ||||+ +|||| |+||+ Sbjct 720 LIGVALLCIWKLLVSFHDRKEVAKFEAERS#KAKWQTGTNPLYRGSTSTFKNVTYK 774 |
| Pongo pygmaeus | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 ||||| |||||||+ ||| # ||| ||+|| | +| | | | Sbjct 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 |
| Felis catus | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 ||||| |||||||+ ||| # ||| ||+|| | +| | | | Sbjct 742 LIGLALLLIWKLLMIIHDTREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 |
| Biomphalaria glabrata | Query 732 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 784 +|| ||+|||| ||| # ||| ||+||+||||| | || Sbjct 733 VGLFLLLLWKLLTFIHDTREFAKFEKERQ#NAKWDTGENPIYKQATSTFKNPTY 785 |
| Xenopus tropicalis | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 ||||| |||||||+ ||| # ||| ||+|| | +| | | | Sbjct 742 LIGLALLLIWKLLMIIHDRREFAKFEKEKM#NAKWDTGENPIYKSAVTTVVNPKYEG 797 |
| Crassostrea gigas | Query 733 GLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYR 785 |+ |||||| || | # ||| ||+||+||||| | ||| Sbjct 746 GIILLLIWKLFTTISDKRELARFEKEAM#NARWDTGENPIYKQATSTFVNPTYR 798 |
| Podocoryne carnea | Query 732 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 784 |||| ||||||| || | # ||+ ||+||+||||| | | Sbjct 726 IGLALLLIWKLLATIQDRREFAKFEKDRQ#NPKWDSGENPIYKKATSTFQNPMY 778 |
| Caenorhabditis briggsae | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 ++|+ ||+|||| +|| # ||| ||+||+||+|| | | | Sbjct 752 ILGILLLLLWKLLTVLHDRAEYAKFNNERL#MAKWDTNENPIYKQATTTFKNPVYAG 807 |
| Caenorhabditis elegans | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 ++|+ ||+|||| +|| # ||| ||+||+||+|| | | | Sbjct 751 ILGILLLLLWKLLTVLHDRSEYATFNNERL#MAKWDTNENPIYKQATTTFKNPVYAG 806 |
| Ictalurus punctatus | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 ||||| | |||||+ ||| # || ||+|| | +| | | | Sbjct 751 LIGLALLFIWKLLMIIHDRREFAKFEKEKM#NAKWDAGENPIYKSAVTTVVNPKYEG 806 |
| Strongylocentrotus purpuratus | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 784 |+||| ||||+|| ||| # |+ ||+|| +|| | | || Sbjct 750 LVGLALLLIWRLLTYIHDRREFQNFEKERA#NATWEGGENPIYKPSTSVFKNPTY 803 |
| Lytechinus variegatus | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 784 |+||| ||||+|| ||| # |+ ||+|| +|| | | || Sbjct 750 LVGLALLLIWRLLTYIHDRREFQNFEKERA#NATWEGGENPIYKPSTSVFKNPTY 803 |
| Pacifastacus leniusculus | Query 732 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTN 781 ||| |||||||| |+|| # | ||||| | +|| | Sbjct 745 IGLLTLLIWKLLTTLHDRREYAKFEKERK#LPSGKRAENPLYKSAKTTFQN 794 |
| Apis mellifera | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXX-WDTANNPLYKEATSTFTNITY 784 +|| | || ||+| | | # |+ +||||||||||| | |+ Sbjct 759 IIGFAILLTWKILTMIRDNREFKKFERERI#LANKWNRRDNPLYKEATSTFKNPTF 813 |
| Manduca sexta | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 784 |||| ||+ ||+|+ +|| # +| + ||||+| |+| | Sbjct 708 LIGLLTLIAWKILVDLHDKREYAKFEEESR#SRGFDVSLNPLYQEPEINFSNPVY 761 |
| Spodoptera frugiperda | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 784 |||+ |++|||+|+ +|| # +| + ||||++ + |+| | Sbjct 723 LIGILTVIIWKILVDMHDKKEYKKFQDEAL#AAGYDVSLNPLYQDPSINFSNPVY 776 |
| Halocynthia roretzi | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 |||+ |+|||++ |+ | # | ||+| |+| | | + | Sbjct 778 LIGIIALIIWKVIQTLRDKREYEKFKKEED#LRKWTKGENPVYVNASSKFDNPMFGG 833 |
| Mus sp. | Query 732 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRGT 787 +|| +| ++| + |+| # | |||||| | +| | ++|| Sbjct 737 VGLGLVLAYRLSVEIYDRREYRRFEKEQQ#QLNWKQDNNPLYKSAITTTVNPRFQGT 792 |
| Schistosoma japonicum | Query 733 GLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITY 784 || |||+||+||| | # |+ | ||+++ |+ | |+ Sbjct 232 GLILLLIYKLVITIDDRRELANFKQQGE#NMRWEMAENPIFESPTTNVLNPTF 283 |
| Oncorhynchus mykiss | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 |||| ||| | ++ + | # | +|||+ ||+| | |+ | Sbjct 721 LIGLLILLIIKAILYMRDVKEFRRFENEQK#KGKWSPGDNPLFMNATTTVANPTFTG 776 |
| Acropora millepora | Query 732 IGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTNITYRG 786 +|| |++ | | |+ | # | ||||+ | +|| | || | Sbjct 735 LGLLILIVIKGLFTMVDRIEYQKFERERM#HSKWTREKNPLYQAAKTTFENPTYAG 789 |
| Bubalus bubalis | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTN 781 |+|+ |+||| | + | # |+ +|||+| ||+| | Sbjct 714 LVGILLLVIWKALTHLSDLREYHRFEKEKL#KSQWNN-DNPLFKSATTTVMN 763 |
| Capra hircus | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTN 781 |+|+ |+||| | + | # |+ +|||+| ||+| | Sbjct 715 LVGILLLVIWKALTHLSDLREYHRFEKEKL#KSQWNN-DNPLFKSATTTVMN 764 |
| Sigmodon hispidus | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTN 781 |||+ |+||| | + | # |+ +|||+| ||+| | Sbjct 714 LIGVLLLVIWKALTHLTDLREYRHFEKEKL#KSQWNN-DNPLFKSATTTVMN 763 |
| Ovis canadensis | Query 731 LIGLATLLIWKLLITIHDXXXXXXXXXXXX#XXXWDTANNPLYKEATSTFTN 781 |+|+ | ||| | + | # |+ +|||+| ||+| | Sbjct 715 LVGILLLAIWKALTHLSDLREYHRFEKEKL#KSQWNN-DNPLFKSATTTVMN 764 |
[Site 2] EERARAKWDT766-ANNPLYKEAT
Thr766  Ala
Ala
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Glu757 | Glu758 | Arg759 | Ala760 | Arg761 | Ala762 | Lys763 | Trp764 | Asp765 | Thr766 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ala767 | Asn768 | Asn769 | Pro770 | Leu771 | Tyr772 | Lys773 | Glu774 | Ala775 | Thr776 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LLIWKLLITIHDRKEFAKFEEERARAKWDTANNPLYKEATSTFTNITYRGT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 74.30 | 21 | - |
| 2 | Mus musculus | 74.30 | 13 | unnamed protein product |
| 3 | Homo sapiens | 74.30 | 12 | integrin beta chain, beta 3 precursor |
| 4 | Bos taurus | 74.30 | 11 | PREDICTED: similar to platelet glycoprotein IIIa i |
| 5 | Macaca mulatta | 74.30 | 10 | PREDICTED: similar to integrin beta chain, beta 3 |
| 6 | Pan troglodytes | 74.30 | 8 | PREDICTED: integrin beta chain, beta 3 isoform 4 |
| 7 | Canis familiaris | 74.30 | 8 | platelet glycoprotein IIIa |
| 8 | Sus scrofa | 74.30 | 5 | AF170527_1 glycoprotein IIIa; GPIIIa |
| 9 | Rattus norvegicus | 74.30 | 5 | integrin beta 3 |
| 10 | synthetic construct | 74.30 | 4 | AF239959_1 hybrid integrin beta 3 subunit precurso |
| 11 | Oryctolagus cuniculus | 74.30 | 2 | glycoprotein IIIa |
| 12 | Equus caballus | 74.30 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 13 | rats, Peptide Partial, 723 aa | 74.30 | 1 | beta 3 integrin, GPIIIA |
| 14 | Canis lupus familiaris | 74.30 | 1 | integrin beta chain, beta 3 |
| 15 | Gallus gallus | 70.90 | 7 | integrin, beta 3 precursor |
| 16 | Monodelphis domestica | 70.10 | 5 | PREDICTED: similar to platelet glycoprotein IIIa |
| 17 | Xenopus laevis | 67.80 | 6 | Unknown (protein for MGC:80757) |
| 18 | mice, Peptide Partial, 680 aa | 65.50 | 1 | beta 3 integrin, GPIIIA |
| 19 | Danio rerio | 64.70 | 10 | hypothetical protein LOC100001836 |
| 20 | Tetraodon nigroviridis | 64.70 | 5 | unnamed protein product |
| 21 | Aedes aegypti | 55.50 | 1 | integrin beta subunit |
| 22 | Tribolium castaneum | 55.10 | 2 | PREDICTED: similar to CG1560-PA |
| 23 | Drosophila melanogaster | 54.30 | 1 | myospheroid CG1560-PA |
| 24 | Ostrinia furnacalis | 53.90 | 1 | integrin beta 1 precursor |
| 25 | Crassostrea gigas | 52.80 | 1 | integrin beta cgh |
| 26 | Anopheles gambiae str. PEST | 52.80 | 1 | ENSANGP00000021373 |
| 27 | Anopheles gambiae | 52.80 | 1 | integrin beta subunit |
| 28 | Biomphalaria glabrata | 52.00 | 1 | beta integrin subunit |
| 29 | Pseudoplusia includens | 50.80 | 1 | integrin beta 1 |
| 30 | Caenorhabditis briggsae | 50.10 | 1 | Hypothetical protein CBG03601 |
| 31 | Caenorhabditis elegans | 50.10 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 32 | Pongo pygmaeus | 49.70 | 2 | hypothetical protein |
| 33 | Ovis aries | 49.70 | 1 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 34 | Podocoryne carnea | 48.10 | 1 | AF308652_1 integrin beta chain |
| 35 | Felis catus | 47.80 | 1 | integrin beta 1 |
| 36 | Xenopus tropicalis | 47.80 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 37 | Ictalurus punctatus | 44.30 | 1 | beta-1 integrin |
| 38 | Strongylocentrotus purpuratus | 43.10 | 4 | integrin beta-C subunit |
| 39 | Lytechinus variegatus | 43.10 | 1 | beta-C integrin subunit |
| 40 | Pacifastacus leniusculus | 39.30 | 1 | integrin |
| 41 | Apis mellifera | 38.90 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 42 | Spodoptera frugiperda | 35.00 | 1 | integrin beta 1 |
| 43 | Manduca sexta | 34.70 | 1 | plasmatocyte-specific integrin beta 1 |
| 44 | Schistosoma japonicum | 33.90 | 1 | SJCHGC06221 protein |
| 45 | Mus sp. | 33.50 | 1 | beta 7 integrin |
| 46 | Halocynthia roretzi | 33.10 | 1 | integrin beta Hr2 precursor |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Mus musculus | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 737 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 787 |
| Homo sapiens | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Bos taurus | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 784 |
| Macaca mulatta | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Pan troglodytes | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Canis familiaris | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 784 |
| Sus scrofa | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 784 |
| Rattus norvegicus | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 737 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 787 |
| synthetic construct | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Oryctolagus cuniculus | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 738 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 788 |
| Equus caballus | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 784 |
| rats, Peptide Partial, 723 aa | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 673 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 723 |
| Canis lupus familiaris | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 |||||||||||| |||#||||||||||||||||||||| Sbjct 734 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYRGT 784 |
| Gallus gallus | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 |||||||||||| |||# ||||||||||||||||||| Sbjct 730 LLIWKLLITIHDRREFARFEEEKARAKWDT#GNNPLYKEATSTFTNITYRG 779 |
| Monodelphis domestica | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYR 785 |||||||||||| |||#||||||||||||||||||| Sbjct 713 LLIWKLLITIHDRREFAKFEEERARAKWDT#ANNPLYKEATSTFTNITYR 761 |
| Xenopus laevis | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 |||||||||||| |||#|+||||| |||||||||||| Sbjct 736 LLIWKLLITIHDRREFAKFEEERAKAKWDT#AHNPLYKGATSTFTNITYRG 785 |
| mice, Peptide Partial, 680 aa | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNIT 783 |||||||||||| |||#||||||||||||||||| Sbjct 634 LLIWKLLITIHDRKEFAKFEEERARAKWDT#ANNPLYKEATSTFTNIT 680 |
| Danio rerio | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 |||||||||||| |+|# +||||| |||||||||||| Sbjct 739 LLIWKLLITIHDRREFAKFEEERARAKWET#GHNPLYKGATSTFTNITYRG 788 |
| Tetraodon nigroviridis | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 |||||||||||| |||# +||||| |||||||||+|| Sbjct 753 LLIWKLLITIHDRREFAKFEEERARAKWDT#GHNPLYKGATSTFTNITFRG 802 |
| Aedes aegypti | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 ||+|||| |||| |||# ||+||+||+|| | || | Sbjct 785 LLLWKLLTTIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAG 834 |
| Tribolium castaneum | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 ||+||| |||| |||# ||+||+||||| | || | Sbjct 779 LLLWKLFTTIHDRREFAKFEKEAMMARWDT#GENPIYKQATSTFKNPTYAG 828 |
| Drosophila melanogaster | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 ||+|||| |||| |||# ||+||+||||| | | | Sbjct 796 LLLWKLLTTIHDRREFARFEKERMNAKWDT#GENPIYKQATSTFKNPMYAG 845 |
| Ostrinia furnacalis | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 |++||++ |||| |||# ||+||+||||| | || | Sbjct 781 LMLWKMVTTIHDRREFARFEKERMMAKWDT#GENPIYKQATSTFKNPTYAG 830 |
| Crassostrea gigas | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYR 785 |||||| || | |||# ||+||+||||| | ||| Sbjct 750 LLIWKLFTTISDKRELARFEKEAMNARWDT#GENPIYKQATSTFVNPTYR 798 |
| Anopheles gambiae str. PEST | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 ||+||+| +||| |||# ||+||+||+|| | || | Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAG 836 |
| Anopheles gambiae | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 ||+||+| +||| |||# ||+||+||+|| | || | Sbjct 787 LLLWKVLTSIHDRREFARFEKERMMAKWDT#GENPIYKQATTTFKNPTYAG 836 |
| Biomphalaria glabrata | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 784 ||+|||| ||| |||# ||+||+||||| | || Sbjct 738 LLLWKLLTFIHDTREFAKFEKERQNAKWDT#GENPIYKQATSTFKNPTY 785 |
| Pseudoplusia includens | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 |++||+ | || |||# ||+||+||||| | || | Sbjct 787 LMLWKMATTSHDRREFARFEKERMMAKWDT#GENPIYKQATSTFKNPTYAG 836 |
| Caenorhabditis briggsae | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 ||+|||| +|| |||# ||+||+||+|| | | | Sbjct 758 LLLWKLLTVLHDRAEYAKFNNERLMAKWDT#NENPIYKQATTTFKNPVYAG 807 |
| Caenorhabditis elegans | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 ||+|||| +|| |||# ||+||+||+|| | | | Sbjct 757 LLLWKLLTVLHDRSEYATFNNERLMAKWDT#NENPIYKQATTTFKNPVYAG 806 |
| Pongo pygmaeus | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYR 785 | |||||++ || | |# ||||+ +|||| |+||+ Sbjct 654 LCIWKLLVSFHDRKEVAKFEAERSKVKWQT#GTNPLYRGSTSTFKNVTYK 702 |
| Ovis aries | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYR 785 | |||||++ || | |# ||||+ +|||| |+||+ Sbjct 726 LCIWKLLVSFHDRKEVAKFEAERSKAKWQT#GTNPLYRGSTSTFKNVTYK 774 |
| Podocoryne carnea | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 784 ||||||| || | ||+# ||+||+||||| | | Sbjct 731 LLIWKLLATIQDRREFAKFEKDRQNPKWDS#GENPIYKKATSTFQNPMY 778 |
| Felis catus | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 |||||||+ ||| |||# ||+|| | +| | | | Sbjct 748 LLIWKLLMIIHDTREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 |
| Xenopus tropicalis | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 |||||||+ ||| |||# ||+|| | +| | | | Sbjct 748 LLIWKLLMIIHDRREFAKFEKEKMNAKWDT#GENPIYKSAVTTVVNPKYEG 797 |
| Ictalurus punctatus | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 | |||||+ ||| || # ||+|| | +| | | | Sbjct 757 LFIWKLLMIIHDRREFAKFEKEKMNAKWDA#GENPIYKSAVTTVVNPKYEG 806 |
| Strongylocentrotus purpuratus | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 784 ||||+|| ||| |+ # ||+|| +|| | | || Sbjct 756 LLIWRLLTYIHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTY 803 |
| Lytechinus variegatus | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 784 ||||+|| ||| |+ # ||+|| +|| | | || Sbjct 756 LLIWRLLTYIHDRREFQNFEKERANATWEG#GENPIYKPSTSVFKNPTY 803 |
| Pacifastacus leniusculus | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTN 781 ||||||| |+|| #| ||||| | +|| | Sbjct 750 LLIWKLLTTLHDRREYAKFEKERKLPSGKR#AENPLYKSAKTTFQN 794 |
| Apis mellifera | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXX-WDT#ANNPLYKEATSTFTNITY 784 || ||+| | | |+ # +||||||||||| | |+ Sbjct 765 LLTWKILTMIRDNREFKKFERERILANKWNR#RDNPLYKEATSTFKNPTF 813 |
| Spodoptera frugiperda | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 784 ++|||+|+ +|| +| #+ ||||++ + |+| | Sbjct 729 VIIWKILVDMHDKKEYKKFQDEALAAGYDV#SLNPLYQDPSINFSNPVY 776 |
| Manduca sexta | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 784 |+ ||+|+ +|| +| #+ ||||+| |+| | Sbjct 714 LIAWKILVDLHDKREYAKFEEESRSRGFDV#SLNPLYQEPEINFSNPVY 761 |
| Schistosoma japonicum | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITY 784 |||+||+||| | |+ #| ||+++ |+ | |+ Sbjct 236 LLIYKLVITIDDRRELANFKQQGENMRWEM#AENPIFESPTTNVLNPTF 283 |
| Mus sp. | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRGT 787 +| ++| + |+| | # |||||| | +| | ++|| Sbjct 742 VLAYRLSVEIYDRREYRRFEKEQQQLNWKQ#DNNPLYKSAITTTVNPRFQGT 792 |
| Halocynthia roretzi | Query 737 LLIWKLLITIHDXXXXXXXXXXXXXXXWDT#ANNPLYKEATSTFTNITYRG 786 |+|||++ |+ | | # ||+| |+| | | + | Sbjct 784 LIIWKVIQTLRDKREYEKFKKEEDLRKWTK#GENPVYVNASSKFDNPMFGG 833 |
[Site 3] ATSTFTNITY784-RGT
Tyr784  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala775 | Thr776 | Ser777 | Thr778 | Phe779 | Thr780 | Asn781 | Ile782 | Thr783 | Tyr784 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg785 | Gly786 | Thr787 | - | - | - | - | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| FEEERARAKWDTANNPLYKEATSTFTNITYRGT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 72.80 | 22 | - |
| 2 | Mus musculus | 72.80 | 13 | integrin beta 3 |
| 3 | Homo sapiens | 72.80 | 12 | integrin beta chain, beta 3 precursor |
| 4 | Bos taurus | 72.80 | 11 | PREDICTED: similar to platelet glycoprotein IIIa i |
| 5 | Macaca mulatta | 72.80 | 10 | PREDICTED: similar to integrin beta chain, beta 3 |
| 6 | Pan troglodytes | 72.80 | 8 | PREDICTED: integrin beta chain, beta 3 isoform 4 |
| 7 | Canis familiaris | 72.80 | 8 | platelet glycoprotein IIIa |
| 8 | Rattus norvegicus | 72.80 | 8 | integrin beta 3 |
| 9 | Sus scrofa | 72.80 | 5 | glycoprotein GPIIIa |
| 10 | synthetic construct | 72.80 | 4 | AF239959_1 hybrid integrin beta 3 subunit precurso |
| 11 | Oryctolagus cuniculus | 72.80 | 2 | glycoprotein IIIa |
| 12 | Equus caballus | 72.80 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 13 | rats, Peptide Partial, 723 aa | 72.80 | 1 | beta 3 integrin, GPIIIA |
| 14 | Canis lupus familiaris | 72.80 | 1 | integrin beta chain, beta 3 |
| 15 | Monodelphis domestica | 68.60 | 5 | PREDICTED: similar to platelet glycoprotein IIIa |
| 16 | Gallus gallus | 68.20 | 7 | integrin, beta 3 precursor |
| 17 | Xenopus laevis | 65.10 | 6 | Unknown (protein for MGC:80757) |
| 18 | mice, Peptide Partial, 680 aa | 63.90 | 1 | beta 3 integrin, GPIIIA |
| 19 | Tetraodon nigroviridis | 63.50 | 5 | unnamed protein product |
| 20 | Danio rerio | 63.20 | 8 | hypothetical protein LOC100001836 |
| 21 | Pseudoplusia includens | 52.00 | 1 | integrin beta 1 |
| 22 | Ostrinia furnacalis | 52.00 | 1 | integrin beta 1 precursor |
| 23 | Anopheles gambiae str. PEST | 50.80 | 1 | ENSANGP00000021373 |
| 24 | Anopheles gambiae | 50.80 | 1 | integrin beta subunit |
| 25 | Aedes aegypti | 50.80 | 1 | integrin beta subunit |
| 26 | Biomphalaria glabrata | 50.40 | 1 | beta integrin subunit |
| 27 | Drosophila melanogaster | 50.10 | 1 | myospheroid CG1560-PA |
| 28 | Crassostrea gigas | 49.30 | 1 | integrin beta cgh |
| 29 | Ovis aries | 48.90 | 1 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 30 | Tribolium castaneum | 48.50 | 2 | PREDICTED: similar to CG1560-PA |
| 31 | Pongo pygmaeus | 47.40 | 2 | hypothetical protein |
| 32 | Caenorhabditis briggsae | 45.80 | 1 | Hypothetical protein CBG03601 |
| 33 | Caenorhabditis elegans | 45.80 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 34 | Podocoryne carnea | 43.10 | 1 | AF308652_1 integrin beta chain |
| 35 | Felis catus | 43.10 | 1 | integrin beta 1 |
| 36 | Xenopus tropicalis | 43.10 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 37 | Strongylocentrotus purpuratus | 41.60 | 4 | integrin beta-C subunit |
| 38 | Lytechinus variegatus | 41.60 | 1 | beta-C integrin subunit |
| 39 | Ictalurus punctatus | 41.20 | 2 | beta-1 integrin |
| 40 | Apis mellifera | 40.80 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 41 | Acropora millepora | 40.00 | 1 | integrin subunit betaCn1 |
| 42 | Oncorhynchus mykiss | 36.60 | 1 | CD18 protein |
| 43 | Mus sp. | 36.60 | 1 | beta 7 integrin |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| Mus musculus | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| Homo sapiens | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| Bos taurus | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| Macaca mulatta | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| Pan troglodytes | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| Canis familiaris | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| Rattus norvegicus | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| Sus scrofa | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| synthetic construct | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| Oryctolagus cuniculus | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| Equus caballus | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| rats, Peptide Partial, 723 aa | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| Canis lupus familiaris | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||||||||||||||||||||||||||||||#||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 |
| Monodelphis domestica | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#R 785 ||||||||||||||||||||||||||||||#| Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#R 785 |
| Gallus gallus | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 ||||+||||||| |||||||||||||||||#|| Sbjct 748 FEEEKARAKWDTGNNPLYKEATSTFTNITY#RG 779 |
| Xenopus laevis | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 ||||||+||||||+||||| ||||||||||#|| Sbjct 754 FEEERAKAKWDTAHNPLYKGATSTFTNITY#RG 785 |
| mice, Peptide Partial, 680 aa | Query 755 FEEERARAKWDTANNPLYKEATSTFTNIT 783 ||||||||||||||||||||||||||||| Query 755 FEEERARAKWDTANNPLYKEATSTFTNIT 783 |
| Tetraodon nigroviridis | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 |||||||||||| |||||| |||||||| |#+| Sbjct 720 FEEERARAKWDTGNNPLYKGATSTFTNIAY#KG 751 |
| Danio rerio | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 ||||||||||+| +||||| ||||||||||#|| Sbjct 757 FEEERARAKWETGHNPLYKGATSTFTNITY#RG 788 |
| Pseudoplusia includens | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 ||+|| ||||| ||+||+||||| | ||# | Sbjct 805 FEKERMMAKWDTGENPIYKQATSTFKNPTY#AG 836 |
| Ostrinia furnacalis | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 ||+|| ||||| ||+||+||||| | ||# | Sbjct 799 FEKERMMAKWDTGENPIYKQATSTFKNPTY#AG 830 |
| Anopheles gambiae str. PEST | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 ||+|| ||||| ||+||+||+|| | ||# | Sbjct 805 FEKERMMAKWDTGENPIYKQATTTFKNPTY#AG 836 |
| Anopheles gambiae | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 ||+|| ||||| ||+||+||+|| | ||# | Sbjct 805 FEKERMMAKWDTGENPIYKQATTTFKNPTY#AG 836 |
| Aedes aegypti | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 ||+|| ||||| ||+||+||+|| | ||# | Sbjct 803 FEKERMMAKWDTGENPIYKQATTTFKNPTY#AG 834 |
| Biomphalaria glabrata | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY# 784 ||+|| ||||| ||+||+||||| | ||# Sbjct 756 FEKERQNAKWDTGENPIYKQATSTFKNPTY# 785 |
| Drosophila melanogaster | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 ||+|| ||||| ||+||+||||| | |# | Sbjct 814 FEKERMNAKWDTGENPIYKQATSTFKNPMY#AG 845 |
| Crassostrea gigas | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#R 785 ||+| |+||| ||+||+||||| | ||#| Sbjct 768 FEKEAMNARWDTGENPIYKQATSTFVNPTY#R 798 |
| Ovis aries | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#R 785 || ||++||| | ||||+ +|||| |+||#+ Sbjct 744 FEAERSKAKWQTGTNPLYRGSTSTFKNVTY#K 774 |
| Tribolium castaneum | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 ||+| |+||| ||+||+||||| | ||# | Sbjct 797 FEKEAMMARWDTGENPIYKQATSTFKNPTY#AG 828 |
| Pongo pygmaeus | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#R 785 || ||++ || | ||||+ +|||| |+||#+ Sbjct 672 FEAERSKVKWQTGTNPLYRGSTSTFKNVTY#K 702 |
| Caenorhabditis briggsae | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 | || ||||| ||+||+||+|| | |# | Sbjct 776 FNNERLMAKWDTNENPIYKQATTTFKNPVY#AG 807 |
| Caenorhabditis elegans | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 | || ||||| ||+||+||+|| | |# | Sbjct 775 FNNERLMAKWDTNENPIYKQATTTFKNPVY#AG 806 |
| Podocoryne carnea | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY# 784 ||++| |||+ ||+||+||||| | |# Sbjct 749 FEKDRQNPKWDSGENPIYKKATSTFQNPMY# 778 |
| Felis catus | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 ||+|+ ||||| ||+|| | +| | |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EG 797 |
| Xenopus tropicalis | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 ||+|+ ||||| ||+|| | +| | |# | Sbjct 766 FEKEKMNAKWDTGENPIYKSAVTTVVNPKY#EG 797 |
| Strongylocentrotus purpuratus | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY# 784 ||+||| | |+ ||+|| +|| | | ||# Sbjct 774 FEKERANATWEGGENPIYKPSTSVFKNPTY# 803 |
| Lytechinus variegatus | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY# 784 ||+||| | |+ ||+|| +|| | | ||# Sbjct 774 FEKERANATWEGGENPIYKPSTSVFKNPTY# 803 |
| Ictalurus punctatus | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 ||+|+ |||| ||+|| | +| | |# | Sbjct 775 FEKEKMNAKWDAGENPIYKSAVTTVVNPKY#EG 806 |
| Apis mellifera | Query 755 FEEERARA-KWDTANNPLYKEATSTFTNITY# 784 || || | ||+ +||||||||||| | |+# Sbjct 783 FERERILANKWNRRDNPLYKEATSTFKNPTF# 813 |
| Acropora millepora | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 || || +|| ||||+ | +|| | ||# | Sbjct 758 FERERMHSKWTREKNPLYQAAKTTFENPTY#AG 789 |
| Oncorhynchus mykiss | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RG 786 || |+ + || +|||+ ||+| | |+# | Sbjct 745 FENEQKKGKWSPGDNPLFMNATTTVANPTF#TG 776 |
| Mus sp. | Query 755 FEEERARAKWDTANNPLYKEATSTFTNITY#RGT 787 ||+|+ + | |||||| | +| | +#+|| Sbjct 760 FEKEQQQLNWKQDNNPLYKSAITTTVNPRF#QGT 792 |
[Site 4] KWDTANNPLY772-KEATSTFTNI
Tyr772  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Lys763 | Trp764 | Asp765 | Thr766 | Ala767 | Asn768 | Asn769 | Pro770 | Leu771 | Tyr772 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys773 | Glu774 | Ala775 | Thr776 | Ser777 | Thr778 | Phe779 | Thr780 | Asn781 | Ile782 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| LITIHDRKEFAKFEEERARAKWDTANNPLYKEATSTFTNITYRGT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 62.00 | 12 | - |
| 2 | Homo sapiens | 62.00 | 8 | integrin beta chain, beta 3 precursor |
| 3 | Pan troglodytes | 62.00 | 7 | PREDICTED: integrin beta chain, beta 3 isoform 4 |
| 4 | Macaca mulatta | 62.00 | 7 | PREDICTED: similar to integrin beta chain, beta 3 |
| 5 | Mus musculus | 62.00 | 7 | unnamed protein product |
| 6 | Canis familiaris | 62.00 | 6 | platelet glycoprotein IIIa |
| 7 | Bos taurus | 62.00 | 5 | PREDICTED: similar to platelet glycoprotein IIIa i |
| 8 | Sus scrofa | 62.00 | 5 | glycoprotein GPIIIa |
| 9 | Rattus norvegicus | 62.00 | 4 | integrin beta 3 |
| 10 | synthetic construct | 62.00 | 3 | AF239959_1 hybrid integrin beta 3 subunit precurso |
| 11 | Oryctolagus cuniculus | 62.00 | 2 | glycoprotein IIIa |
| 12 | Canis lupus familiaris | 62.00 | 1 | integrin beta chain, beta 3 |
| 13 | Equus caballus | 62.00 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 14 | rats, Peptide Partial, 723 aa | 62.00 | 1 | beta 3 integrin, GPIIIA |
| 15 | Gallus gallus | 58.50 | 3 | integrin, beta 3 precursor |
| 16 | Monodelphis domestica | 57.80 | 3 | PREDICTED: similar to platelet glycoprotein IIIa |
| 17 | Xenopus laevis | 55.50 | 4 | Unknown (protein for MGC:80757) |
| 18 | mice, Peptide Partial, 680 aa | 53.10 | 1 | beta 3 integrin, GPIIIA |
| 19 | Danio rerio | 52.40 | 8 | hypothetical protein LOC100001836 |
| 20 | Tetraodon nigroviridis | 52.40 | 5 | unnamed protein product |
| 21 | Tribolium castaneum | 43.90 | 2 | PREDICTED: similar to CG1560-PA |
| 22 | Ostrinia furnacalis | 43.90 | 1 | integrin beta 1 precursor |
| 23 | Aedes aegypti | 43.90 | 1 | integrin beta subunit |
| 24 | Drosophila melanogaster | 42.70 | 1 | myospheroid CG1560-PA |
| 25 | Anopheles gambiae | 42.40 | 1 | integrin beta subunit |
| 26 | Anopheles gambiae str. PEST | 42.40 | 1 | ENSANGP00000021373 |
| 27 | Pseudoplusia includens | 41.60 | 1 | integrin beta 1 |
| 28 | Crassostrea gigas | 41.20 | 1 | integrin beta cgh |
| 29 | Biomphalaria glabrata | 40.40 | 1 | beta integrin subunit |
| 30 | Pongo pygmaeus | 39.30 | 2 | hypothetical protein |
| 31 | Ovis aries | 39.30 | 1 | ITB6_SHEEP Integrin beta-6 precursor emb |
| 32 | Caenorhabditis briggsae | 38.50 | 1 | Hypothetical protein CBG03601 |
| 33 | Caenorhabditis elegans | 38.50 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 34 | Podocoryne carnea | 35.80 | 1 | AF308652_1 integrin beta chain |
| 35 | Xenopus tropicalis | 35.40 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 36 | Felis catus | 35.40 | 1 | integrin beta 1 |
| 37 | Apis mellifera | 35.00 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 38 | Strongylocentrotus purpuratus | 34.30 | 2 | integrin beta L subunit |
| 39 | Ictalurus punctatus | 33.50 | 1 | beta-1 integrin |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Homo sapiens | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Pan troglodytes | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Macaca mulatta | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Mus musculus | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 743 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 787 |
| Canis familiaris | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 784 |
| Bos taurus | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 784 |
| Sus scrofa | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 784 |
| Rattus norvegicus | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 743 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 787 |
| synthetic construct | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Oryctolagus cuniculus | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 744 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 788 |
| Canis lupus familiaris | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 784 |
| Equus caballus | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 784 |
| rats, Peptide Partial, 723 aa | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRGT 787 |||||| |||||||||#||||||||||||||| Sbjct 679 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNITYRGT 723 |
| Gallus gallus | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 |||||| ||| |||||#|||||||||||||| Sbjct 736 LITIHDRREFARFEEEKARAKWDTGNNPLY#KEATSTFTNITYRG 779 |
| Monodelphis domestica | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYR 785 |||||| |||||||||#||||||||||||| Sbjct 719 LITIHDRREFAKFEEERARAKWDTANNPLY#KEATSTFTNITYR 761 |
| Xenopus laevis | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 |||||| ||||+||||#| |||||||||||| Sbjct 742 LITIHDRREFAKFEEERAKAKWDTAHNPLY#KGATSTFTNITYRG 785 |
| mice, Peptide Partial, 680 aa | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNIT 783 |||||| |||||||||#||||||||||| Sbjct 640 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTNIT 680 |
| Danio rerio | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 |||||| |+| +||||#| |||||||||||| Sbjct 745 LITIHDRREFAKFEEERARAKWETGHNPLY#KGATSTFTNITYRG 788 |
| Tetraodon nigroviridis | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 |||||| ||| +||||#| |||||||||+|| Sbjct 759 LITIHDRREFAKFEEERARAKWDTGHNPLY#KGATSTFTNITFRG 802 |
| Tribolium castaneum | Query 745 TIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 |||| ||| ||+|#|+||||| | || | Sbjct 787 TIHDRREFAKFEKEAMMARWDTGENPIY#KQATSTFKNPTYAG 828 |
| Ostrinia furnacalis | Query 745 TIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 |||| ||| ||+|#|+||||| | || | Sbjct 789 TIHDRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYAG 830 |
| Aedes aegypti | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 | |||| ||| ||+|#|+||+|| | || | Sbjct 791 LTTIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYAG 834 |
| Drosophila melanogaster | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 | |||| ||| ||+|#|+||||| | | | Sbjct 802 LTTIHDRREFARFEKERMNAKWDTGENPIY#KQATSTFKNPMYAG 845 |
| Anopheles gambiae | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 | +||| ||| ||+|#|+||+|| | || | Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYAG 836 |
| Anopheles gambiae str. PEST | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 | +||| ||| ||+|#|+||+|| | || | Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYAG 836 |
| Pseudoplusia includens | Query 745 TIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 | || ||| ||+|#|+||||| | || | Sbjct 795 TSHDRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYAG 836 |
| Crassostrea gigas | Query 764 WDTANNPLY#KEATSTFTNITYR 785 ||| ||+|#|+||||| | ||| Sbjct 777 WDTGENPIY#KQATSTFVNPTYR 798 |
| Biomphalaria glabrata | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITY 784 | ||| ||| ||+|#|+||||| | || Sbjct 744 LTFIHDTREFAKFEKERQNAKWDTGENPIY#KQATSTFKNPTY 785 |
| Pongo pygmaeus | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYR 785 |++ || | | ||||#+ +|||| |+||+ Sbjct 660 LVSFHDRKEVAKFEAERSKVKWQTGTNPLY#RGSTSTFKNVTYK 702 |
| Ovis aries | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYR 785 |++ || | | ||||#+ +|||| |+||+ Sbjct 732 LVSFHDRKEVAKFEAERSKAKWQTGTNPLY#RGSTSTFKNVTYK 774 |
| Caenorhabditis briggsae | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 | +|| ||| ||+|#|+||+|| | | | Sbjct 764 LTVLHDRAEYAKFNNERLMAKWDTNENPIY#KQATTTFKNPVYAG 807 |
| Caenorhabditis elegans | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 | +|| ||| ||+|#|+||+|| | | | Sbjct 763 LTVLHDRSEYATFNNERLMAKWDTNENPIY#KQATTTFKNPVYAG 806 |
| Podocoryne carnea | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITY 784 | || | ||+ ||+|#|+||||| | | Sbjct 737 LATIQDRREFAKFEKDRQNPKWDSGENPIY#KKATSTFQNPMY 778 |
| Xenopus tropicalis | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 |+ ||| ||| ||+|#| | +| | | | Sbjct 754 LMIIHDRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEG 797 |
| Felis catus | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 |+ ||| ||| ||+|#| | +| | | | Sbjct 754 LMIIHDTREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKYEG 797 |
| Apis mellifera | Query 764 WDTANNPLY#KEATSTFTNITY 784 |+ +||||#||||||| | |+ Sbjct 793 WNRRDNPLY#KEATSTFKNPTF 813 |
| Strongylocentrotus purpuratus | Query 764 WDTANNPLY#KEATSTFTNITY 784 || ++||+|#| +|+|| | || Sbjct 780 WDQSDNPIY#KSSTTTFKNPTY 800 |
| Ictalurus punctatus | Query 743 LITIHDXXXXXXXXXXXXXXXWDTANNPLY#KEATSTFTNITYRG 786 |+ ||| || ||+|#| | +| | | | Sbjct 763 LMIIHDRREFAKFEKEKMNAKWDAGENPIY#KSAVTTVVNPKYEG 806 |
* References
[PubMed ID: 19122172] Guo M, Daines D, Tang J, Shen Q, Perrin RM, Takada Y, Yuan SY, Wu MH, Fibrinogen-gamma C-terminal fragments induce endothelial barrier dysfunction and microvascular leak via integrin-mediated and RhoA-dependent mechanism. Arterioscler Thromb Vasc Biol. 2009 Mar;29(3):394-400. Epub 2009 Jan 2.
[PubMed ID: 19027743] ... Srinivasan KR, Blesson CS, Fatima I, Kitchlu S, Jain SK, Mehrotra PK, Dwivedi A, Expression of alphaVbeta3 integrin in rat endometrial epithelial cells and its functional role during implantation. Gen Comp Endocrinol. 2009 Jan 15;160(2):124-33. Epub 2008 Nov 6.
[PubMed ID: 18638458] ... Kang WS, Choi JS, Shin YJ, Kim HY, Cha JH, Lee JY, Chun MH, Lee MY, Differential regulation of osteopontin receptors, CD44 and the alpha(v) and beta(3) integrin subunits, in the rat hippocampus following transient forebrain ischemia. Brain Res. 2008 Sep 4;1228:208-16. Epub 2008 Jul 3.
[PubMed ID: 18256073] ... Higuchi T, Bengel FM, Seidl S, Watzlowik P, Kessler H, Hegenloh R, Reder S, Nekolla SG, Wester HJ, Schwaiger M, Assessment of alphavbeta3 integrin expression after myocardial infarction by positron emission tomography. Cardiovasc Res. 2008 May 1;78(2):395-403. Epub 2008 Feb 6.
[PubMed ID: 17868879] ... Han M, Wen JK, Zheng B, Liu Z, Chen Y, Blockade of integrin beta3-FAK signaling pathway activated by osteopontin inhibits neointimal formation after balloon injury. Cardiovasc Pathol. 2007 Sep-Oct;16(5):283-90. Epub 2007 Jun 20.
[PubMed ID: 15044441] ... Zhou X, Murphy FR, Gehdu N, Zhang J, Iredale JP, Benyon RC, Engagement of alphavbeta3 integrin regulates proliferation and apoptosis of hepatic stellate cells. J Biol Chem. 2004 Jun 4;279(23):23996-4006. Epub 2004 Mar 24.
[PubMed ID: 12639933] ... Campbell S, Otis M, Cote M, Gallo-Payet N, Payet MD, Connection between integrins and cell activation in rat adrenal glomerulosa cells: a role for Arg-Gly-Asp peptide in the activation of the p42/p44(mapk) pathway and intracellular calcium. Endocrinology. 2003 Apr;144(4):1486-95.
[PubMed ID: 12606526] ... Kanzaki T, Otabe M, Latent transforming growth factor-beta binding protein-1, a component of latent transforming growth factor-beta complex, accelerates the migration of aortic smooth muscle cells in diabetic rats through integrin-beta3. Diabetes. 2003 Mar;52(3):824-8.
[PubMed ID: 12600920] ... Sun M, Opavsky MA, Stewart DJ, Rabinovitch M, Dawood F, Wen WH, Liu PP, Temporal response and localization of integrins beta1 and beta3 in the heart after myocardial infarction: regulation by cytokines. Circulation. 2003 Feb 25;107(7):1046-52.
[PubMed ID: 11891802] ... Simovich MJ, Conrad ME, Umbreit JN, Moore EG, Hainsworth LN, Smith HK, Cellular location of proteins related to iron absorption and transport. Am J Hematol. 2002 Mar;69(3):164-70.