XSB0961 : integrin beta 2 [Mus musculus]
[ CaMP Format ]
This entry is computationally expanded from SB0002
* Basic Information
| Organism | Mus musculus (house mouse) |
| Protein Names | Integrin beta-2; Cell surface adhesion glycoproteins LFA-1/CR3/p150,95 subunit beta; Complement receptor C3 subunit beta; integrin beta 2; Mac-1 beta; macrophage antigen-1 beta; lymphocyte function associated antigen 1 |
| Gene Names | Itgb2; integrin beta 2 |
| Gene Locus | 10 41.5 cM; chromosome 10 |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 770 aa
Average Mass: 84.894 kDa
Monoisotopic Mass: 84.838 kDa
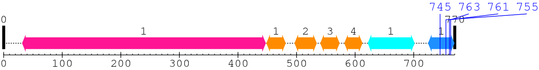
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Integrin_beta 1. | 32 | 447 | 1064.1 | 0 |
| EGF_2 1. | 449 | 481 | 20.8 | 0.0058 |
| EGF_2 2. | 497 | 534 | 16.1 | 0.041 |
| EGF_2 3. | 541 | 573 | 27.1 | 7e-05 |
| EGF_2 4. | 582 | 612 | 26.5 | 0.00011 |
| Integrin_B_tail 1. | 622 | 701 | 121.7 | 2.4e-33 |
| Integrin_b_cyt 1. | 725 | 769 | 85.5 | 1.9e-22 |
| --- cleavage 745 (inside Integrin_b_cyt 725..769) --- | ||||
| --- cleavage 763 (inside Integrin_b_cyt 725..769) --- | ||||
| --- cleavage 761 (inside Integrin_b_cyt 725..769) --- | ||||
| --- cleavage 755 (inside Integrin_b_cyt 725..769) --- | ||||
3. Sequence Information
Fasta Sequence: XSB0961.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
4 [sites]
Cleavage sites (±10aa)
[Site 1] YRRFEKEKLK745-SQWNNDNPLF
Lys745  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Tyr736 | Arg737 | Arg738 | Phe739 | Glu740 | Lys741 | Glu742 | Lys743 | Leu744 | Lys745 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser746 | Gln747 | Trp748 | Asn749 | Asn750 | Asp751 | Asn752 | Pro753 | Leu754 | Phe755 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| IGVLLLVIWKALTHLTDLREYRRFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 115.00 | 29 | I |
| 2 | Mus musculus | 115.00 | 16 | integrin beta 2 |
| 3 | Rattus norvegicus | 113.00 | 10 | PREDICTED: similar to Integrin beta-2 precursor (C |
| 4 | Sigmodon hispidus | 113.00 | 1 | AF445415_1 integrin beta-2 precursor |
| 5 | Homo sapiens | 110.00 | 25 | leukocyte adhesion protein beta-subunit precursor |
| 6 | Pan troglodytes | 110.00 | 14 | integrin, beta 2 |
| 7 | Bos taurus | 110.00 | 13 | integrin, beta 2 |
| 8 | synthetic construct | 110.00 | 5 | integrin, beta 2 (antigen CD18 (p95), lymphocyte f |
| 9 | Bubalus bubalis | 110.00 | 1 | integrin beta-2 |
| 10 | Capra hircus | 110.00 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 11 | Ovis aries | 109.00 | 2 | antigen CD18 |
| 12 | Ovis canadensis | 109.00 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 13 | Sus scrofa | 107.00 | 6 | CD18 leukocyte adhesion molecule |
| 14 | Canis familiaris | 106.00 | 10 | AF181965_1 B-2 integrin |
| 15 | Monodelphis domestica | 89.40 | 6 | PREDICTED: similar to Stromal interaction molecule |
| 16 | Gallus gallus | 80.90 | 7 | integrin, beta 2 |
| 17 | Xenopus laevis | 67.00 | 6 | hypothetical protein LOC379708 |
| 18 | Felis catus | 64.70 | 1 | integrin beta 1 |
| 19 | Danio rerio | 64.30 | 14 | integrin, beta 1b |
| 20 | Tetraodon nigroviridis | 64.30 | 8 | unnamed protein product |
| 21 | Oryctolagus cuniculus | 64.30 | 2 | integrin beta 1 |
| 22 | Pongo pygmaeus | 64.30 | 2 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 23 | Xenopus tropicalis | 64.30 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 24 | Pacifastacus leniusculus | 63.90 | 1 | integrin |
| 25 | Ictalurus punctatus | 63.50 | 2 | beta-1 integrin |
| 26 | Oncorhynchus mykiss | 63.50 | 1 | CD18 protein |
| 27 | Anopheles gambiae str. PEST | 62.40 | 1 | ENSANGP00000021373 |
| 28 | Anopheles gambiae | 62.40 | 1 | integrin beta subunit |
| 29 | Caenorhabditis briggsae | 62.00 | 1 | Hypothetical protein CBG03601 |
| 30 | Aedes aegypti | 61.60 | 1 | integrin beta subunit |
| 31 | Cyprinus carpio | 60.80 | 2 | CD18 type 2 |
| 32 | Caenorhabditis elegans | 60.80 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 33 | Drosophila melanogaster | 60.50 | 1 | myospheroid CG1560-PA |
| 34 | Ostrinia furnacalis | 59.70 | 1 | integrin beta 1 precursor |
| 35 | Crassostrea gigas | 59.30 | 1 | integrin beta cgh |
| 36 | Tribolium castaneum | 58.90 | 3 | PREDICTED: similar to CG1560-PA |
| 37 | Biomphalaria glabrata | 58.20 | 1 | beta integrin subunit |
| 38 | Apis mellifera | 57.40 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 39 | Pseudoplusia includens | 57.40 | 1 | integrin beta 1 |
| 40 | Strongylocentrotus purpuratus | 55.80 | 6 | PREDICTED: similar to integrin beta L subunit, par |
| 41 | Podocoryne carnea | 55.50 | 1 | AF308652_1 integrin beta chain |
| 42 | Mus sp. | 55.10 | 1 | beta 7 integrin |
| 43 | Macaca mulatta | 54.70 | 13 | PREDICTED: integrin, beta 7 |
| 44 | Lytechinus variegatus | 53.90 | 1 | beta-C integrin subunit |
| 45 | Acropora millepora | 52.00 | 1 | integrin subunit betaCn1 |
| 46 | Canis lupus familiaris | 49.30 | 1 | integrin beta chain, beta 3 |
| 47 | rats, Peptide Partial, 723 aa | 49.30 | 1 | beta 3 integrin, GPIIIA |
| 48 | mice, Peptide Partial, 680 aa | 49.30 | 1 | beta 3 integrin, GPIIIA |
| 49 | Equus caballus | 49.30 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 50 | Halocynthia roretzi | 48.50 | 2 | integrin beta Hr2 precursor |
| 51 | Schistosoma japonicum | 45.80 | 1 | SJCHGC06221 protein |
| 52 | Ophlitaspongia tenuis | 44.70 | 1 | integrin subunit betaPo1 |
| 53 | Geodia cydonium | 43.10 | 1 | integrin beta subunit |
| 54 | Spodoptera frugiperda | 42.00 | 1 | integrin beta 1 |
| 55 | Suberites domuncula | 39.70 | 1 | integrin beta subunit |
| 56 | Manduca sexta | 39.70 | 1 | plasmatocyte-specific integrin beta 1 |
| 57 | Drosophila pseudoobscura | 33.90 | 1 | GA14581-PA |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |
| Mus musculus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||| Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |
| Rattus norvegicus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |||||||||||||||||| |||||||||||#||||||||||||||||||||||||| Sbjct 714 IGVLLLVIWKALTHLTDLNEYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 768 |
| Sigmodon hispidus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |||||||||||||||||||||| |||||||#||||||||||||||||||||||||| Sbjct 715 IGVLLLVIWKALTHLTDLREYRHFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| Homo sapiens | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 ||+||||||||| ||+||||||||||||||#||||||||||||||||||||||||| Sbjct 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| Pan troglodytes | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 ||+||||||||| ||+||||||||||||||#||||||||||||||||||||||||| Sbjct 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| Bos taurus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 +|+||||||||||||+||||| ||||||||#||||||||||||||||||||||||| Sbjct 715 VGILLLVIWKALTHLSDLREYHRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| synthetic construct | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 ||+||||||||| ||+||||||||||||||#||||||||||||||||||||||||| Sbjct 715 IGILLLVIWKALIHLSDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| Bubalus bubalis | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 +|+||||||||||||+||||| ||||||||#||||||||||||||||||||||||| Sbjct 715 VGILLLVIWKALTHLSDLREYHRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 769 |
| Capra hircus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 +|+||||||||||||+||||| ||||||||#||||||||||||||||||||||||| Sbjct 716 VGILLLVIWKALTHLSDLREYHRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |
| Ovis aries | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 +|+||| ||||||||+||||| ||||||||#||||||||||||||||||||||||| Sbjct 716 VGILLLAIWKALTHLSDLREYHRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |
| Ovis canadensis | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 +|+||| ||||||||+||||| ||||||||#||||||||||||||||||||||||| Sbjct 716 VGILLLAIWKALTHLSDLREYHRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 |
| Sus scrofa | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAE 769 +|+ |||||| ||||+|||||+||||||||#|||||||||||||||||||||||| Sbjct 715 VGIFLLVIWKVLTHLSDLREYKRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAE 768 |
| Canis familiaris | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFAES 770 ||+||| ||||||||+||||++|||||||+#||||||||||||||||||||+|||| Sbjct 710 IGILLLAIWKALTHLSDLREFKRFEKEKLR#SQWNNDNPLFKSATTTVMNPRFAES 764 |
| Monodelphis domestica | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNP 765 |||||| +|| ||| +|||||+||||||||#||||||||||| |||++ | Sbjct 1181 IGVLLLAVWKVLTHFSDLREYKRFEKEKLK#SQWNNDNPLFKKKTTTIIIP 1230 |
| Gallus gallus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKF 767 ||+|||+ |+ || + | |||||||||| |#++|| |||||||||||||||+| Sbjct 717 IGLLLLLTWRLLTEIFDRREYRRFEKEKSK#AKWNEADNPLFKSATTTVMNPRF 769 |
| Xenopus laevis | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 768 ||+ |+||| +| + | ||+|||||++ #|+|| |||+ +||||| || |+ Sbjct 717 IGLACLIIWKIITEIKDRNEYQRFEKERMN#SKWNPGHNPLYHNATTTVQNPNFS 770 |
| Felis catus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 767 ||+ ||+||| | + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 743 IGLALLLIWKLLMIIHDTREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 795 |
| Danio rerio | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 767 ||+ ||+||| | + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 740 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDAGENPIYKSAVTTVVNPKY 792 |
| Tetraodon nigroviridis | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 767 ||+ ||+||| | + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 736 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 788 |
| Oryctolagus cuniculus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 767 ||+ ||+||| | + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 8 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 60 |
| Pongo pygmaeus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 767 ||+ ||+||| | + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 795 |
| Xenopus tropicalis | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 767 ||+ ||+||| | + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 743 IGLALLLIWKLLMIIHDRREFAKFEKEKMN#AKWDTGENPIYKSAVTTVVNPKY 795 |
| Pacifastacus leniusculus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKE-KLK#SQWNNDNPLFKSATTTVMNPKFAES 770 ||+| |+||| || | | ||| +|||| || #| +|||+||| || || ||+| Sbjct 745 IGLLTLLIWKLLTTLHDRREYAKFEKERKLP#SGKRAENPLYKSAKTTFQNPAFAQS 800 |
| Ictalurus punctatus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 767 ||+ || ||| | + | ||+ +|||||+ #++|+ +||++||| |||+|||+ Sbjct 752 IGLALLFIWKLLMIIHDRREFAKFEKEKMN#AKWDAGENPIYKSAVTTVVNPKY 804 |
| Oncorhynchus mykiss | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 767 ||+|+|+| ||+ ++ |++|+|||| |+ |# +|+ ||||| +||||| || | Sbjct 722 IGLLILLIIKAILYMRDVKEFRRFENEQKK#GKWSPGDNPLFMNATTTVANPTF 774 |
| Anopheles gambiae str. PEST | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 768 ||+ +|++|| || + | ||+ |||||++ #++|+ +||++| |||| || +| Sbjct 782 IGMAVLLLWKVLTSIHDRREFARFEKERMM#AKWDTGENPIYKQATTTFKNPTYA 835 |
| Anopheles gambiae | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 768 ||+ +|++|| || + | ||+ |||||++ #++|+ +||++| |||| || +| Sbjct 782 IGMAVLLLWKVLTSIHDRREFARFEKERMM#AKWDTGENPIYKQATTTFKNPTYA 835 |
| Caenorhabditis briggsae | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 768 +|+|||++|| || | | || +| |+| #++|+ |+||++| |||| || +| Sbjct 753 LGILLLLLWKLLTVLHDRAEYAKFNNERLM#AKWDTNENPIYKQATTTFKNPVYA 806 |
| Aedes aegypti | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 768 ||+ +|++|| || + | ||+ |||||++ #++|+ +||++| |||| || +| Sbjct 780 IGLAVLLLWKLLTTIHDRREFARFEKERMM#AKWDTGENPIYKQATTTFKNPTYA 833 |
| Cyprinus carpio | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPKFA 768 ||+|+|+| ||+ + +||||++||||++ # + + ||||++||||| || |+ Sbjct 712 IGLLILLIIKAVLYASDLREWKRFEKDRKH#EKTSGTNPLFQNATTTVQNPTFS 764 |
| Caenorhabditis elegans | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 768 +|+|||++|| || | | || | |+| #++|+ |+||++| |||| || +| Sbjct 752 LGILLLLLWKLLTVLHDRSEYATFNNERLM#AKWDTNENPIYKQATTTFKNPVYA 805 |
| Drosophila melanogaster | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 768 +|+ +|++|| || + | ||+ |||||++ #++|+ +||++| ||+| || +| Sbjct 791 VGLAILLLWKLLTTIHDRREFARFEKERMN#AKWDTGENPIYKQATSTFKNPMYA 844 |
| Ostrinia furnacalis | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 768 +|+ ||++|| +| + | ||+ |||||++ #++|+ +||++| ||+| || +| Sbjct 776 VGLALLMLWKMVTTIHDRREFARFEKERMM#AKWDTGENPIYKQATSTFKNPTYA 829 |
| Crassostrea gigas | Query 717 GVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFAE 769 |++||+||| | ++| || ||||| + #++|+ +||++| ||+| +|| + + Sbjct 746 GIILLLIWKLFTTISDKRELARFEKEAMN#ARWDTGENPIYKQATSTFVNPTYRQ 799 |
| Tribolium castaneum | Query 717 GVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 767 ||+||+ || | + | +|| +|| |+ |# ||+ |||||++ ||+| || + Sbjct 700 GVILLLTWKVCTTVHDRQEYAKFENERQK#MQWHRNDNPLYRQATSTFKNPVY 751 |
| Biomphalaria glabrata | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 767 +|+ ||++|| || + | ||+ +||||+ #++|+ +||++| ||+| || + Sbjct 733 VGLFLLLLWKLLTFIHDTREFAKFEKERQN#AKWDTGENPIYKQATSTFKNPTY 785 |
| Apis mellifera | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEK-LK#SQWN-NDNPLFKSATTTVMNPKF 767 || +|+ || || + | ||+++||+|+ | #++|| ||||+| ||+| || | Sbjct 760 IGFAILLTWKILTMIRDNREFKKFERERILA#NKWNRRDNPLYKEATSTFKNPTF 813 |
| Pseudoplusia includens | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFA 768 +|+ ||++|| | | ||+ |||||++ #++|+ +||++| ||+| || +| Sbjct 782 VGLALLMLWKMATTSHDRREFARFEKERMM#AKWDTGENPIYKQATSTFKNPTYA 835 |
| Strongylocentrotus purpuratus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKFAE 769 ||++|+ |+ |++ | ||| ++| + |#+||+ +|||++||+||| || + + Sbjct 94 IGMILVCAWRLYTYVQDKREYAQWENDCKK#AQWDQSDNPIYKSSTTTFKNPTYGK 148 |
| Podocoryne carnea | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKF 767 ||+ ||+||| | + | ||+ +|||++ # +|++ +||++| ||+| || + Sbjct 726 IGLALLLIWKLLATIQDRREFAKFEKDRQN#PKWDSGENPIYKKATSTFQNPMY 778 |
| Mus sp. | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNND-NPLFKSATTTVMNPKF 767 +|+ |++ ++ + | |||||||||+ +# | | |||+||| || +||+| Sbjct 737 VGLGLVLAYRLSVEIYDRREYRRFEKEQQQ#LNWKQDNNPLYKSAITTTVNPRF 789 |
| Macaca mulatta | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNND-NPLFKSATTTVMNPKFAES 770 +|+ |++ ++ + | ||| |||||+ +# | | |||+||| || +||+| |+ Sbjct 614 VGLGLVLAYRLSVEIYDRREYSRFEKEQQQ#LNWKQDSNPLYKSAITTTINPRFQEA 669 |
| Lytechinus variegatus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQW-NNDNPLFKSATTTVMNPKF 767 +|+ ||+||+ ||++ | ||++ ||||+ #+ | +||++| +|+ || + Sbjct 751 VGLALLLIWRLLTYIHDRREFQNFEKERAN#ATWEGGENPIYKPSTSVFKNPTY 803 |
| Acropora millepora | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNND-NPLFKSATTTVMNPKFA 768 +|+|+|++ | | + | ||++||+|++ #|+| + |||+++| || || +| Sbjct 735 LGLLILIVIKGLFTMVDRIEYQKFERERMH#SKWTREKNPLYQAAKTTFENPTYA 788 |
| Canis lupus familiaris | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 764 ||+ |+||| | + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 729 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 778 |
| rats, Peptide Partial, 723 aa | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 764 ||+ |+||| | + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 668 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 717 |
| mice, Peptide Partial, 680 aa | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 764 ||+ |+||| | + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 629 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 678 |
| Equus caballus | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMN 764 ||+ |+||| | + | +|+ +||+|+ +#++|+ +|||+| ||+| | Sbjct 729 IGLATLLIWKLLITIHDRKEFAKFEEERAR#AKWDTANNPLYKEATSTFTN 778 |
| Halocynthia roretzi | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQW-NNDNPLFKSATTTVMNPKF 767 ||++ |+||| + | | ||| +|+||+ # +| +||++ +|++ || | Sbjct 779 IGIIALIIWKVIQTLRDKREYEKFKKEEDL#RKWTKGENPVYVNASSKFDNPMF 831 |
| Schistosoma japonicum | Query 717 GVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNN-DNPLFKSATTTVMNPKFAES 770 |++||+|+| + + | || |+++ # +| +||+|+| || |+|| | |+ Sbjct 232 GLILLLIYKLVITIDDRRELANFKQQGEN#MRWEMAENPIFESPTTNVLNPTFEEN 286 |
| Ophlitaspongia tenuis | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQW-NNDNPLFKSATTTVMNPKFAE 769 +|+|||++ | | | |+ | |+||+| #+++ |+|||++||| || + + Sbjct 784 LGLLLLLLLKGLLLLWDVVEVRKFEREIKN#AKYTKNENPLYRSATKDYQNPLYGK 838 |
| Geodia cydonium | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQW-NNDNPLFKSATTTVMNPKFAE 769 +|+| |++ | | + | | ++|| | |# ++ |+|||+ |||| || + + Sbjct 824 LGILALILLKILLMVLDYTELKKFENELAK#VKYTKNENPLYHSATTEHKNPIYGQ 878 |
| Spodoptera frugiperda | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 767 ||+| ++||| | + | +||++|+ | | #+ ++ + |||++ + || + Sbjct 724 IGILTVIIWKILVDMHDKKEYKKFQDEALA#AGYDVSLNPLYQDPSINFSNPVY 776 |
| Suberites domuncula | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQW-NNDNPLFKSATTTVMNPKFAE 769 +||+ |++ | + |+ | ++|||| |#+++ + |||++|| || + + Sbjct 710 LGVIALLLLKLFLFIIDVIEVKKFEKELAK#TKYPKHQNPLYRSAAKDYQNPIYGQ 764 |
| Manduca sexta | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWN-NDNPLFKSATTTVMNPKF 767 ||+| |+ || | | | ||| +||+| # ++ + |||++ || + Sbjct 709 IGLLTLIAWKILVDLHDKREYAKFEEESRS#RGFDVSLNPLYQEPEINFSNPVY 761 |
| Drosophila pseudoobscura | Query 716 IGVLLLVIWKALTHLTDLREYRRFEKEKLK#SQWNNDNPLFKSATTTVMNPK 766 ||+||+ ++ | ||| |||+|+ |# + + +|||++ + || Sbjct 710 IGLLLIFLFIWCIRRKDAREYARFEEEQAK#-RVSQENPLYRDPVGRYVVPK 759 |
[Site 2] LFKSATTTVM763-NPKFAES
Met763  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Leu754 | Phe755 | Lys756 | Ser757 | Ala758 | Thr759 | Thr760 | Thr761 | Val762 | Met763 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn764 | Pro765 | Lys766 | Phe767 | Ala768 | Glu769 | Ser770 | - | - | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| REYRRFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 80.50 | 20 | A |
| 2 | Homo sapiens | 80.50 | 14 | integrin, beta 2 precursor |
| 3 | Mus musculus | 80.50 | 10 | integrin beta 2 |
| 4 | Pan troglodytes | 80.50 | 6 | integrin, beta 2 |
| 5 | synthetic construct | 80.50 | 3 | integrin, beta 2 (antigen CD18 (p95), lymphocyte f |
| 6 | Rattus norvegicus | 78.60 | 8 | PREDICTED: similar to Integrin beta-2 precursor (C |
| 7 | Bos taurus | 78.60 | 6 | integrin, beta 2 |
| 8 | Ovis canadensis | 78.60 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 9 | Sigmodon hispidus | 78.60 | 1 | AF445415_1 integrin beta-2 precursor |
| 10 | Capra hircus | 78.60 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 11 | Ovis aries | 78.60 | 1 | antigen CD18 |
| 12 | Bubalus bubalis | 78.60 | 1 | integrin beta-2 |
| 13 | Sus scrofa | 77.80 | 4 | CD18 leukocyte adhesion molecule |
| 14 | Canis familiaris | 75.50 | 4 | AF181965_1 B-2 integrin |
| 15 | Gallus gallus | 62.40 | 5 | integrin, beta 2 |
| 16 | Monodelphis domestica | 61.20 | 4 | PREDICTED: similar to Stromal interaction molecule |
| 17 | Xenopus laevis | 48.90 | 5 | hypothetical protein LOC379708 |
| 18 | Mus sp. | 48.90 | 1 | beta 7 integrin |
| 19 | Macaca mulatta | 48.50 | 2 | PREDICTED: integrin, beta 7 |
| 20 | Danio rerio | 48.10 | 12 | integrin, beta 1b |
| 21 | Tetraodon nigroviridis | 48.10 | 5 | unnamed protein product |
| 22 | Oryctolagus cuniculus | 48.10 | 2 | integrin beta 1 |
| 23 | Ictalurus punctatus | 48.10 | 2 | beta-1 integrin |
| 24 | Xenopus tropicalis | 48.10 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 25 | Felis catus | 48.10 | 1 | integrin beta 1 |
| 26 | Pongo pygmaeus | 48.10 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 27 | Oncorhynchus mykiss | 47.00 | 1 | CD18 protein |
| 28 | Anopheles gambiae | 44.70 | 1 | integrin beta subunit |
| 29 | Anopheles gambiae str. PEST | 44.70 | 1 | ENSANGP00000021373 |
| 30 | Aedes aegypti | 44.70 | 1 | integrin beta subunit |
| 31 | Pacifastacus leniusculus | 44.30 | 1 | integrin |
| 32 | Cyprinus carpio | 43.90 | 2 | CD18 type 2 |
| 33 | Drosophila melanogaster | 43.50 | 1 | myospheroid CG1560-PA |
| 34 | Tribolium castaneum | 43.10 | 2 | PREDICTED: similar to CG1560-PA |
| 35 | Pseudoplusia includens | 43.10 | 1 | integrin beta 1 |
| 36 | Ostrinia furnacalis | 43.10 | 1 | integrin beta 1 precursor |
| 37 | Caenorhabditis briggsae | 42.00 | 1 | Hypothetical protein CBG03601 |
| 38 | Apis mellifera | 42.00 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 39 | Strongylocentrotus purpuratus | 41.20 | 5 | integrin beta L subunit |
| 40 | Caenorhabditis elegans | 40.80 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 41 | Crassostrea gigas | 40.40 | 1 | integrin beta cgh |
| 42 | Acropora millepora | 40.40 | 1 | integrin subunit betaCn1 |
| 43 | Biomphalaria glabrata | 39.30 | 1 | beta integrin subunit |
| 44 | Podocoryne carnea | 38.10 | 1 | AF308652_1 integrin beta chain |
| 45 | Lytechinus variegatus | 35.00 | 1 | beta-C integrin subunit |
| 46 | mice, Peptide Partial, 680 aa | 34.70 | 1 | beta 3 integrin, GPIIIA |
| 47 | rats, Peptide Partial, 723 aa | 34.70 | 1 | beta 3 integrin, GPIIIA |
| 48 | Canis lupus familiaris | 34.70 | 1 | integrin beta chain, beta 3 |
| 49 | Equus caballus | 34.70 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 50 | Schistosoma japonicum | 33.90 | 1 | SJCHGC06221 protein |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 ||||||||||||||||||||||||||||||#||||||| Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Homo sapiens | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 ||||||||||||||||||||||||||||||#||||||| Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Mus musculus | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 ||||||||||||||||||||||||||||||#||||||| Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Pan troglodytes | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 ||||||||||||||||||||||||||||||#||||||| Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| synthetic construct | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 ||||||||||||||||||||||||||||||#||||||| Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Rattus norvegicus | Query 735 EYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |||||||||||||||||||||||||||||#||||||| Query 735 EYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Bos taurus | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 ||| ||||||||||||||||||||||||||#||||||| Sbjct 733 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Ovis canadensis | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 ||| ||||||||||||||||||||||||||#||||||| Sbjct 734 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Sigmodon hispidus | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |||| |||||||||||||||||||||||||#||||||| Sbjct 733 REYRHFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Capra hircus | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 ||| ||||||||||||||||||||||||||#||||||| Sbjct 734 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Ovis aries | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 ||| ||||||||||||||||||||||||||#||||||| Sbjct 734 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 |
| Bubalus bubalis | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 ||| ||||||||||||||||||||||||||#||||||| Sbjct 733 REYHRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 769 |
| Sus scrofa | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAE 769 |||+||||||||||||||||||||||||||#|||||| Sbjct 733 REYKRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAE 768 |
| Canis familiaris | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFAES 770 ||++|||||||+||||||||||||||||||#||+|||| Sbjct 728 REFKRFEKEKLRSQWNNDNPLFKSATTTVM#NPRFAES 764 |
| Gallus gallus | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 |||||||||| |++|| |||||||||||||#||+| Sbjct 735 REYRRFEKEKSKAKWNEADNPLFKSATTTVM#NPRF 769 |
| Monodelphis domestica | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NP 765 |||+||||||||||||||||||| |||++# | Sbjct 1199 REYKRFEKEKLKSQWNNDNPLFKKKTTTII#IP 1230 |
| Xenopus laevis | Query 735 EYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 768 ||+|||||++ |+|| |||+ +||||| #|| |+ Sbjct 736 EYQRFEKERMNSKWNPGHNPLYHNATTTVQ#NPNFS 770 |
| Mus sp. | Query 734 REYRRFEKEKLKSQWNND-NPLFKSATTTVM#NPKF 767 |||||||||+ + | | |||+||| || +#||+| Sbjct 755 REYRRFEKEQQQLNWKQDNNPLYKSAITTTV#NPRF 789 |
| Macaca mulatta | Query 734 REYRRFEKEKLKSQWNND-NPLFKSATTTVM#NPKFAES 770 ||| |||||+ + | | |||+||| || +#||+| |+ Sbjct 632 REYSRFEKEQQQLNWKQDSNPLYKSAITTTI#NPRFQEA 669 |
| Danio rerio | Query 734 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 767 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 758 REFAKFEKEKMNAKWDAGENPIYKSAVTTVV#NPKY 792 |
| Tetraodon nigroviridis | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 754 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 788 |
| Oryctolagus cuniculus | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 26 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 60 |
| Ictalurus punctatus | Query 734 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 767 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 770 REFAKFEKEKMNAKWDAGENPIYKSAVTTVV#NPKY 804 |
| Xenopus tropicalis | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |
| Felis catus | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |
| Pongo pygmaeus | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 ||+ +|||||+ ++|+ +||++||| |||+#|||+ Sbjct 761 REFAKFEKEKMNAKWDTGENPIYKSAVTTVV#NPKY 795 |
| Oncorhynchus mykiss | Query 734 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 767 +|+|||| |+ | +|+ ||||| +||||| #|| | Sbjct 740 KEFRRFENEQKKGKWSPGDNPLFMNATTTVA#NPTF 774 |
| Anopheles gambiae | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 768 ||+ |||||++ ++|+ +||++| |||| #|| +| Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYA 835 |
| Anopheles gambiae str. PEST | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 768 ||+ |||||++ ++|+ +||++| |||| #|| +| Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYA 835 |
| Aedes aegypti | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 768 ||+ |||||++ ++|+ +||++| |||| #|| +| Sbjct 798 REFARFEKERMMAKWDTGENPIYKQATTTFK#NPTYA 833 |
| Pacifastacus leniusculus | Query 734 REYRRFEKE-KLKSQWNNDNPLFKSATTTVM#NPKFAES 770 ||| +|||| || | +|||+||| || #|| ||+| Sbjct 763 REYAKFEKERKLPSGKRAENPLYKSAKTTFQ#NPAFAQS 800 |
| Cyprinus carpio | Query 734 REYRRFEKEKLKSQWNNDNPLFKSATTTVM#NPKFA 768 ||++||||++ + + ||||++||||| #|| |+ Sbjct 730 REWKRFEKDRKHEKTSGTNPLFQNATTTVQ#NPTFS 764 |
| Drosophila melanogaster | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 768 ||+ |||||++ ++|+ +||++| ||+| #|| +| Sbjct 809 REFARFEKERMNAKWDTGENPIYKQATSTFK#NPMYA 844 |
| Tribolium castaneum | Query 734 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKF 767 +|| +|| |+ | ||+ |||||++ ||+| #|| + Sbjct 717 QEYAKFENERQKMQWHRNDNPLYRQATSTFK#NPVY 751 |
| Pseudoplusia includens | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 768 ||+ |||||++ ++|+ +||++| ||+| #|| +| Sbjct 800 REFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYA 835 |
| Ostrinia furnacalis | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFA 768 ||+ |||||++ ++|+ +||++| ||+| #|| +| Sbjct 794 REFARFEKERMMAKWDTGENPIYKQATSTFK#NPTYA 829 |
| Caenorhabditis briggsae | Query 735 EYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 768 || +| |+| ++|+ |+||++| |||| #|| +| Sbjct 772 EYAKFNNERLMAKWDTNENPIYKQATTTFK#NPVYA 806 |
| Apis mellifera | Query 734 REYRRFEKEK-LKSQWNN-DNPLFKSATTTVM#NPKF 767 ||+++||+|+ | ++|| ||||+| ||+| #|| | Sbjct 778 REFKKFERERILANKWNRRDNPLYKEATSTFK#NPTF 813 |
| Strongylocentrotus purpuratus | Query 734 REYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFAE 769 ||| ++| + |+||+ +|||++||+||| #|| + + Sbjct 766 REYAQWENDCKKAQWDQSDNPIYKSSTTTFK#NPTYGK 802 |
| Caenorhabditis elegans | Query 735 EYRRFEKEKLKSQWN-NDNPLFKSATTTVM#NPKFA 768 || | |+| ++|+ |+||++| |||| #|| +| Sbjct 771 EYATFNNERLMAKWDTNENPIYKQATTTFK#NPVYA 805 |
| Crassostrea gigas | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFAE 769 || ||||| + ++|+ +||++| ||+| +#|| + + Sbjct 763 RELARFEKEAMNARWDTGENPIYKQATSTFV#NPTYRQ 799 |
| Acropora millepora | Query 735 EYRRFEKEKLKSQWNND-NPLFKSATTTVM#NPKFA 768 ||++||+|++ |+| + |||+++| || #|| +| Sbjct 754 EYQKFERERMHSKWTREKNPLYQAAKTTFE#NPTYA 788 |
| Biomphalaria glabrata | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 ||+ +||||+ ++|+ +||++| ||+| #|| + Sbjct 751 REFAKFEKERQNAKWDTGENPIYKQATSTFK#NPTY 785 |
| Podocoryne carnea | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKF 767 ||+ +|||++ +|++ +||++| ||+| #|| + Sbjct 744 REFAKFEKDRQNPKWDSGENPIYKKATSTFQ#NPMY 778 |
| Lytechinus variegatus | Query 734 REYRRFEKEKLKSQW-NNDNPLFKSATTTVM#NPKF 767 ||++ ||||+ + | +||++| +|+ #|| + Sbjct 769 REFQNFEKERANATWEGGENPIYKPSTSVFK#NPTY 803 |
| mice, Peptide Partial, 680 aa | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 764 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 647 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 678 |
| rats, Peptide Partial, 723 aa | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 764 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 686 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 717 |
| Canis lupus familiaris | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 764 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 778 |
| Equus caballus | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#N 764 +|+ +||+|+ +++|+ +|||+| ||+| #| Sbjct 747 KEFAKFEEERARAKWDTANNPLYKEATSTFT#N 778 |
| Schistosoma japonicum | Query 734 REYRRFEKEKLKSQWNN-DNPLFKSATTTVM#NPKFAES 770 || |+++ +| +||+|+| || |+#|| | |+ Sbjct 249 RELANFKQQGENMRWEMAENPIFESPTTNVL#NPTFEEN 286 |
[Site 3] NPLFKSATTT761-VMNPKFAES
Thr761  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Asn752 | Pro753 | Leu754 | Phe755 | Lys756 | Ser757 | Ala758 | Thr759 | Thr760 | Thr761 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val762 | Met763 | Asn764 | Pro765 | Lys766 | Phe767 | Ala768 | Glu769 | Ser770 | - |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DLREYRRFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 84.30 | 20 | A |
| 2 | Homo sapiens | 84.30 | 14 | leukocyte adhesion protein beta-subunit precursor |
| 3 | Mus musculus | 84.30 | 10 | integrin beta 2 |
| 4 | Pan troglodytes | 84.30 | 6 | integrin, beta 2 |
| 5 | synthetic construct | 84.30 | 3 | integrin, beta 2 (antigen CD18 (p95), lymphocyte f |
| 6 | Rattus norvegicus | 82.40 | 8 | PREDICTED: similar to Integrin beta-2 precursor (C |
| 7 | Bos taurus | 82.40 | 6 | integrin, beta 2 |
| 8 | Sigmodon hispidus | 82.40 | 1 | AF445415_1 integrin beta-2 precursor |
| 9 | Capra hircus | 82.40 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 10 | Bubalus bubalis | 82.40 | 1 | integrin beta-2 |
| 11 | Ovis aries | 82.40 | 1 | antigen CD18 |
| 12 | Ovis canadensis | 82.40 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 13 | Sus scrofa | 81.60 | 4 | CD18 leukocyte adhesion molecule |
| 14 | Canis familiaris | 79.30 | 4 | AF181965_1 B-2 integrin |
| 15 | Monodelphis domestica | 65.10 | 4 | PREDICTED: similar to Stromal interaction molecule |
| 16 | Gallus gallus | 63.90 | 5 | integrin, beta 2 |
| 17 | Xenopus laevis | 50.40 | 5 | hypothetical protein LOC379708 |
| 18 | Mus sp. | 50.40 | 1 | beta 7 integrin |
| 19 | Macaca mulatta | 50.10 | 2 | PREDICTED: integrin, beta 7 |
| 20 | Felis catus | 50.10 | 1 | integrin beta 1 |
| 21 | Danio rerio | 49.70 | 12 | integrin, beta 1b |
| 22 | Tetraodon nigroviridis | 49.70 | 6 | unnamed protein product |
| 23 | Oryctolagus cuniculus | 49.70 | 2 | integrin beta 1 |
| 24 | Ictalurus punctatus | 49.70 | 2 | beta-1 integrin |
| 25 | Oncorhynchus mykiss | 49.70 | 1 | CD18 protein |
| 26 | Pongo pygmaeus | 49.70 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 27 | Xenopus tropicalis | 49.70 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 28 | Cyprinus carpio | 47.80 | 2 | CD18 type 2 |
| 29 | Anopheles gambiae str. PEST | 46.20 | 1 | ENSANGP00000021373 |
| 30 | Anopheles gambiae | 46.20 | 1 | integrin beta subunit |
| 31 | Aedes aegypti | 46.20 | 1 | integrin beta subunit |
| 32 | Pacifastacus leniusculus | 45.80 | 1 | integrin |
| 33 | Drosophila melanogaster | 45.10 | 1 | myospheroid CG1560-PA |
| 34 | Tribolium castaneum | 44.70 | 3 | PREDICTED: similar to CG1560-PA |
| 35 | Pseudoplusia includens | 44.70 | 1 | integrin beta 1 |
| 36 | Ostrinia furnacalis | 44.70 | 1 | integrin beta 1 precursor |
| 37 | Apis mellifera | 43.10 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 38 | Caenorhabditis briggsae | 43.10 | 1 | Hypothetical protein CBG03601 |
| 39 | Strongylocentrotus purpuratus | 42.70 | 5 | integrin beta L subunit |
| 40 | Caenorhabditis elegans | 42.00 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 41 | Crassostrea gigas | 42.00 | 1 | integrin beta cgh |
| 42 | Biomphalaria glabrata | 41.20 | 1 | beta integrin subunit |
| 43 | Acropora millepora | 40.80 | 1 | integrin subunit betaCn1 |
| 44 | Podocoryne carnea | 39.70 | 1 | AF308652_1 integrin beta chain |
| 45 | Lytechinus variegatus | 36.60 | 1 | beta-C integrin subunit |
| 46 | Equus caballus | 36.20 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 47 | Canis lupus familiaris | 36.20 | 1 | integrin beta chain, beta 3 |
| 48 | rats, Peptide Partial, 723 aa | 36.20 | 1 | beta 3 integrin, GPIIIA |
| 49 | mice, Peptide Partial, 680 aa | 36.20 | 1 | beta 3 integrin, GPIIIA |
| 50 | Schistosoma japonicum | 35.40 | 1 | SJCHGC06221 protein |
| 51 | Ophlitaspongia tenuis | 33.50 | 1 | integrin subunit betaPo1 |
| 52 | Geodia cydonium | 33.10 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 ||||||||||||||||||||||||||||||#||||||||| Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Homo sapiens | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 ||||||||||||||||||||||||||||||#||||||||| Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Mus musculus | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 ||||||||||||||||||||||||||||||#||||||||| Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Pan troglodytes | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 ||||||||||||||||||||||||||||||#||||||||| Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| synthetic construct | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 ||||||||||||||||||||||||||||||#||||||||| Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Rattus norvegicus | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 || |||||||||||||||||||||||||||#||||||||| Sbjct 730 DLNEYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 768 |
| Bos taurus | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 ||||| ||||||||||||||||||||||||#||||||||| Sbjct 731 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Sigmodon hispidus | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |||||| |||||||||||||||||||||||#||||||||| Sbjct 731 DLREYRHFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Capra hircus | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 ||||| ||||||||||||||||||||||||#||||||||| Sbjct 732 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Bubalus bubalis | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 ||||| ||||||||||||||||||||||||#||||||||| Sbjct 731 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 769 |
| Ovis aries | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 ||||| ||||||||||||||||||||||||#||||||||| Sbjct 732 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Ovis canadensis | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 ||||| ||||||||||||||||||||||||#||||||||| Sbjct 732 DLREYHRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 |
| Sus scrofa | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAE 769 |||||+||||||||||||||||||||||||#|||||||| Sbjct 731 DLREYKRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAE 768 |
| Canis familiaris | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFAES 770 ||||++|||||||+||||||||||||||||#||||+|||| Sbjct 726 DLREFKRFEKEKLRSQWNNDNPLFKSATTT#VMNPRFAES 764 |
| Monodelphis domestica | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNP 765 |||||+||||||||||||||||||| |||#++ | Sbjct 1197 DLREYKRFEKEKLKSQWNNDNPLFKKKTTT#IIIP 1230 |
| Gallus gallus | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 767 | |||||||||| |++|| |||||||||||#||||+| Sbjct 733 DRREYRRFEKEKSKAKWNEADNPLFKSATTT#VMNPRF 769 |
| Xenopus laevis | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 768 | ||+|||||++ |+|| |||+ +||||#| || |+ Sbjct 733 DRNEYQRFEKERMNSKWNPGHNPLYHNATTT#VQNPNFS 770 |
| Mus sp. | Query 732 DLREYRRFEKEKLKSQWNND-NPLFKSATTT#VMNPKF 767 | |||||||||+ + | | |||+||| ||# +||+| Sbjct 753 DRREYRRFEKEQQQLNWKQDNNPLYKSAITT#TVNPRF 789 |
| Macaca mulatta | Query 732 DLREYRRFEKEKLKSQWNND-NPLFKSATTT#VMNPKFAES 770 | ||| |||||+ + | | |||+||| ||# +||+| |+ Sbjct 630 DRREYSRFEKEQQQLNWKQDSNPLYKSAITT#TINPRFQEA 669 |
| Felis catus | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 767 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 759 DTREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |
| Danio rerio | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 767 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 756 DRREFAKFEKEKMNAKWDAGENPIYKSAVTT#VVNPKY 792 |
| Tetraodon nigroviridis | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 767 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 752 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 788 |
| Oryctolagus cuniculus | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 767 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 24 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 60 |
| Ictalurus punctatus | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 767 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 768 DRREFAKFEKEKMNAKWDAGENPIYKSAVTT#VVNPKY 804 |
| Oncorhynchus mykiss | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 767 |++|+|||| |+ | +|+ ||||| +||||#| || | Sbjct 738 DVKEFRRFENEQKKGKWSPGDNPLFMNATTT#VANPTF 774 |
| Pongo pygmaeus | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 767 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |
| Xenopus tropicalis | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 767 | ||+ +|||||+ ++|+ +||++||| ||#|+|||+ Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIYKSAVTT#VVNPKY 795 |
| Cyprinus carpio | Query 732 DLREYRRFEKEKLKSQWNNDNPLFKSATTT#VMNPKFA 768 ||||++||||++ + + ||||++||||#| || |+ Sbjct 728 DLREWKRFEKDRKHEKTSGTNPLFQNATTT#VQNPTFS 764 |
| Anopheles gambiae str. PEST | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 768 | ||+ |||||++ ++|+ +||++| ||||# || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYA 835 |
| Anopheles gambiae | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 768 | ||+ |||||++ ++|+ +||++| ||||# || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYA 835 |
| Aedes aegypti | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFA 768 | ||+ |||||++ ++|+ +||++| ||||# || +| Sbjct 796 DRREFARFEKERMMAKWDTGENPIYKQATTT#FKNPTYA 833 |
| Pacifastacus leniusculus | Query 732 DLREYRRFEKE-KLKSQWNNDNPLFKSATTT#VMNPKFAES 770 | ||| +|||| || | +|||+||| ||# || ||+| Sbjct 761 DRREYAKFEKERKLPSGKRAENPLYKSAKTT#FQNPAFAQS 800 |
| Drosophila melanogaster | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 768 | ||+ |||||++ ++|+ +||++| ||+|# || +| Sbjct 807 DRREFARFEKERMNAKWDTGENPIYKQATST#FKNPMYA 844 |
| Tribolium castaneum | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 767 | +|| +|| |+ | ||+ |||||++ ||+|# || + Sbjct 715 DRQEYAKFENERQKMQWHRNDNPLYRQATST#FKNPVY 751 |
| Pseudoplusia includens | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 768 | ||+ |||||++ ++|+ +||++| ||+|# || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYA 835 |
| Ostrinia furnacalis | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 768 | ||+ |||||++ ++|+ +||++| ||+|# || +| Sbjct 792 DRREFARFEKERMMAKWDTGENPIYKQATST#FKNPTYA 829 |
| Apis mellifera | Query 732 DLREYRRFEKEK-LKSQWNN-DNPLFKSATTT#VMNPKF 767 | ||+++||+|+ | ++|| ||||+| ||+|# || | Sbjct 776 DNREFKKFERERILANKWNRRDNPLYKEATST#FKNPTF 813 |
| Caenorhabditis briggsae | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 768 | || +| |+| ++|+ |+||++| ||||# || +| Sbjct 769 DRAEYAKFNNERLMAKWDTNENPIYKQATTT#FKNPVYA 806 |
| Strongylocentrotus purpuratus | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFAE 769 | ||| ++| + |+||+ +|||++||+|||# || + + Sbjct 764 DKREYAQWENDCKKAQWDQSDNPIYKSSTTT#FKNPTYGK 802 |
| Caenorhabditis elegans | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFA 768 | || | |+| ++|+ |+||++| ||||# || +| Sbjct 768 DRSEYATFNNERLMAKWDTNENPIYKQATTT#FKNPVYA 805 |
| Crassostrea gigas | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFAE 769 | || ||||| + ++|+ +||++| ||+|# +|| + + Sbjct 761 DKRELARFEKEAMNARWDTGENPIYKQATST#FVNPTYRQ 799 |
| Biomphalaria glabrata | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKF 767 | ||+ +||||+ ++|+ +||++| ||+|# || + Sbjct 749 DTREFAKFEKERQNAKWDTGENPIYKQATST#FKNPTY 785 |
| Acropora millepora | Query 732 DLREYRRFEKEKLKSQWNND-NPLFKSATTT#VMNPKFA 768 | ||++||+|++ |+| + |||+++| ||# || +| Sbjct 751 DRIEYQKFERERMHSKWTREKNPLYQAAKTT#FENPTYA 788 |
| Podocoryne carnea | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKF 767 | ||+ +|||++ +|++ +||++| ||+|# || + Sbjct 742 DRREFAKFEKDRQNPKWDSGENPIYKKATST#FQNPMY 778 |
| Lytechinus variegatus | Query 732 DLREYRRFEKEKLKSQW-NNDNPLFKSATTT#VMNPKF 767 | ||++ ||||+ + | +||++| +|+ # || + Sbjct 767 DRREFQNFEKERANATWEGGENPIYKPSTSV#FKNPTY 803 |
| Equus caballus | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 764 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 745 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 778 |
| Canis lupus familiaris | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 764 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 745 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 778 |
| rats, Peptide Partial, 723 aa | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 764 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 684 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 717 |
| mice, Peptide Partial, 680 aa | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMN 764 | +|+ +||+|+ +++|+ +|||+| ||+|# | Sbjct 645 DRKEFAKFEEERARAKWDTANNPLYKEATST#FTN 678 |
| Schistosoma japonicum | Query 732 DLREYRRFEKEKLKSQWNN-DNPLFKSATTT#VMNPKFAES 770 | || |+++ +| +||+|+| || #|+|| | |+ Sbjct 247 DRRELANFKQQGENMRWEMAENPIFESPTTN#VLNPTFEEN 286 |
| Ophlitaspongia tenuis | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFAE 769 |+ | |+||+| +++ |+|||++||| # || + + Sbjct 800 DVVEVRKFEREIKNAKYTKNENPLYRSATKD#YQNPLYGK 838 |
| Geodia cydonium | Query 732 DLREYRRFEKEKLKSQWN-NDNPLFKSATTT#VMNPKFAE 769 | | ++|| | | ++ |+|||+ |||| # || + + Sbjct 840 DYTELKKFENELAKVKYTKNENPLYHSATTE#HKNPIYGQ 878 |
[Site 4] SQWNNDNPLF755-KSATTTVMNP
Phe755  Lys
Lys
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser746 | Gln747 | Trp748 | Asn749 | Asn750 | Asp751 | Asn752 | Pro753 | Leu754 | Phe755 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Lys756 | Ser757 | Ala758 | Thr759 | Thr760 | Thr761 | Val762 | Met763 | Asn764 | Pro765 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ALTHLTDLREYRRFEKEKLKSQWNNDNPLFKSATTTVMNPKFAES |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | N/A | 95.90 | 20 | I |
| 2 | Mus musculus | 95.90 | 10 | integrin beta 2 |
| 3 | Rattus norvegicus | 94.00 | 8 | PREDICTED: similar to Integrin beta-2 precursor (C |
| 4 | Sigmodon hispidus | 94.00 | 1 | AF445415_1 integrin beta-2 precursor |
| 5 | Bos taurus | 92.40 | 6 | integrin, beta 2 |
| 6 | Ovis aries | 92.40 | 1 | antigen CD18 |
| 7 | Ovis canadensis | 92.40 | 1 | ITB2_OVICA Integrin beta-2 precursor (Cell surface |
| 8 | Bubalus bubalis | 92.40 | 1 | integrin beta-2 |
| 9 | Capra hircus | 92.40 | 1 | ITB2_CAPHI Integrin beta-2 precursor (Cell surface |
| 10 | Homo sapiens | 92.00 | 14 | integrin beta chain, beta 2 precursor variant |
| 11 | Pan troglodytes | 92.00 | 6 | integrin, beta 2 |
| 12 | synthetic construct | 92.00 | 3 | integrin, beta 2 (antigen CD18 (p95), lymphocyte f |
| 13 | Sus scrofa | 90.10 | 4 | CD18 leukocyte adhesion molecule |
| 14 | Canis familiaris | 89.40 | 4 | AF181965_1 B-2 integrin |
| 15 | Monodelphis domestica | 72.00 | 4 | PREDICTED: similar to Stromal interaction molecule |
| 16 | Gallus gallus | 67.40 | 6 | integrin, beta 2 |
| 17 | Xenopus laevis | 53.50 | 5 | hypothetical protein LOC379708 |
| 18 | Oncorhynchus mykiss | 52.80 | 1 | CD18 protein |
| 19 | Ictalurus punctatus | 50.80 | 2 | CD18 |
| 20 | Mus sp. | 50.40 | 1 | beta 7 integrin |
| 21 | Cyprinus carpio | 50.10 | 2 | CD18 type 2 |
| 22 | Macaca mulatta | 50.10 | 2 | PREDICTED: integrin, beta 7 |
| 23 | Felis catus | 50.10 | 1 | integrin beta 1 |
| 24 | Danio rerio | 49.70 | 12 | integrin, beta 1b |
| 25 | Tetraodon nigroviridis | 49.70 | 6 | unnamed protein product |
| 26 | Oryctolagus cuniculus | 49.70 | 2 | integrin beta 1 |
| 27 | Pongo pygmaeus | 49.70 | 1 | ITB1_PONPY Integrin beta-1 precursor (Fibronectin |
| 28 | Xenopus tropicalis | 49.70 | 1 | integrin beta 1 (fibronectin receptor beta) |
| 29 | Pacifastacus leniusculus | 49.30 | 1 | integrin |
| 30 | Anopheles gambiae str. PEST | 49.30 | 1 | ENSANGP00000021373 |
| 31 | Anopheles gambiae | 49.30 | 1 | integrin beta subunit |
| 32 | Aedes aegypti | 48.90 | 1 | integrin beta subunit |
| 33 | Drosophila melanogaster | 47.80 | 1 | myospheroid CG1560-PA |
| 34 | Apis mellifera | 46.20 | 1 | PREDICTED: similar to myospheroid CG1560-PA |
| 35 | Caenorhabditis briggsae | 46.20 | 1 | Hypothetical protein CBG03601 |
| 36 | Ostrinia furnacalis | 46.20 | 1 | integrin beta 1 precursor |
| 37 | Strongylocentrotus purpuratus | 45.40 | 5 | integrin beta L subunit |
| 38 | Tribolium castaneum | 45.40 | 3 | PREDICTED: similar to CG1560-PA |
| 39 | Caenorhabditis elegans | 45.10 | 1 | Paralysed Arrest at Two-fold family member (pat-3) |
| 40 | Pseudoplusia includens | 44.70 | 1 | integrin beta 1 |
| 41 | Crassostrea gigas | 44.30 | 1 | integrin beta cgh |
| 42 | Biomphalaria glabrata | 44.30 | 1 | beta integrin subunit |
| 43 | Acropora millepora | 41.60 | 1 | integrin subunit betaCn1 |
| 44 | Podocoryne carnea | 40.80 | 1 | AF308652_1 integrin beta chain |
| 45 | Lytechinus variegatus | 40.80 | 1 | beta-C integrin subunit |
| 46 | Canis lupus familiaris | 36.60 | 1 | integrin beta chain, beta 3 |
| 47 | mice, Peptide Partial, 680 aa | 36.60 | 1 | beta 3 integrin, GPIIIA |
| 48 | rats, Peptide Partial, 723 aa | 36.60 | 1 | beta 3 integrin, GPIIIA |
| 49 | Equus caballus | 36.60 | 1 | integrin, beta 3 (platelet glycoprotein IIIa, anti |
| 50 | Schistosoma japonicum | 35.80 | 1 | SJCHGC06221 protein |
| 51 | Ophlitaspongia tenuis | 34.30 | 1 | integrin subunit betaPo1 |
| 52 | Geodia cydonium | 33.50 | 1 | integrin beta subunit |
Top-ranked sequences
| organism | matching |
|---|---|
| N/A | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 ||||||||||||||||||||||||||||||#||||||||||||||| Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |
| Mus musculus | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 ||||||||||||||||||||||||||||||#||||||||||||||| Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |
| Rattus norvegicus | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |||||||| |||||||||||||||||||||#||||||||||||||| Sbjct 724 ALTHLTDLNEYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 768 |
| Sigmodon hispidus | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |||||||||||| |||||||||||||||||#||||||||||||||| Sbjct 725 ALTHLTDLREYRHFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Bos taurus | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |||||+||||| ||||||||||||||||||#||||||||||||||| Sbjct 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Ovis aries | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |||||+||||| ||||||||||||||||||#||||||||||||||| Sbjct 726 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |
| Ovis canadensis | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |||||+||||| ||||||||||||||||||#||||||||||||||| Sbjct 726 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |
| Bubalus bubalis | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |||||+||||| ||||||||||||||||||#||||||||||||||| Sbjct 725 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Capra hircus | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |||||+||||| ||||||||||||||||||#||||||||||||||| Sbjct 726 ALTHLSDLREYHRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |
| Homo sapiens | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 || ||+||||||||||||||||||||||||#||||||||||||||| Sbjct 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Pan troglodytes | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 || ||+||||||||||||||||||||||||#||||||||||||||| Sbjct 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| synthetic construct | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 || ||+||||||||||||||||||||||||#||||||||||||||| Sbjct 725 ALIHLSDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 769 |
| Sus scrofa | Query 727 LTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAE 769 ||||+|||||+||||||||||||||||||#|||||||||||||| Sbjct 726 LTHLSDLREYKRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAE 768 |
| Canis familiaris | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFAES 770 |||||+||||++|||||||+||||||||||#||||||||||+|||| Sbjct 720 ALTHLSDLREFKRFEKEKLRSQWNNDNPLF#KSATTTVMNPRFAES 764 |
| Monodelphis domestica | Query 727 LTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNP 765 ||| +|||||+||||||||||||||||||#| |||++ | Sbjct 1192 LTHFSDLREYKRFEKEKLKSQWNNDNPLF#KKKTTTIIIP 1230 |
| Gallus gallus | Query 727 LTHLTDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 767 || + | |||||||||| |++|| |||||#||||||||||+| Sbjct 728 LTEIFDRREYRRFEKEKSKAKWNEADNPLF#KSATTTVMNPRF 769 |
| Xenopus laevis | Query 727 LTHLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 768 +| + | ||+|||||++ |+|| |||+# +||||| || |+ Sbjct 728 ITEIKDRNEYQRFEKERMNSKWNPGHNPLY#HNATTTVQNPNFS 770 |
| Oncorhynchus mykiss | Query 726 ALTHLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 767 |+ ++ |++|+|||| |+ | +|+ |||||# +||||| || | Sbjct 732 AILYMRDVKEFRRFENEQKKGKWSPGDNPLF#MNATTTVANPTF 774 |
| Ictalurus punctatus | Query 726 ALTHLTDLREYRRFEKEKLKSQW-NNDNPLF#KSATTTVMNPKF 767 || + ||+|+++|||| + || +||||#++||||| || | Sbjct 726 ALFYFKDLKEWKKFEKEAQRRQWAKGENPLF#QNATTTVANPAF 768 |
| Mus sp. | Query 732 DLREYRRFEKEKLKSQWNND-NPLF#KSATTTVMNPKF 767 | |||||||||+ + | | |||+#||| || +||+| Sbjct 753 DRREYRRFEKEQQQLNWKQDNNPLY#KSAITTTVNPRF 789 |
| Cyprinus carpio | Query 726 ALTHLTDLREYRRFEKEKLKSQWNNDNPLF#KSATTTVMNPKFA 768 |+ + +||||++||||++ + + ||||#++||||| || |+ Sbjct 722 AVLYASDLREWKRFEKDRKHEKTSGTNPLF#QNATTTVQNPTFS 764 |
| Macaca mulatta | Query 732 DLREYRRFEKEKLKSQWNND-NPLF#KSATTTVMNPKFAES 770 | ||| |||||+ + | | |||+#||| || +||+| |+ Sbjct 630 DRREYSRFEKEQQQLNWKQDSNPLY#KSAITTTINPRFQEA 669 |
| Felis catus | Query 732 DLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 767 | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 759 DTREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 795 |
| Danio rerio | Query 732 DLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 767 | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 756 DRREFAKFEKEKMNAKWDAGENPIY#KSAVTTVVNPKY 792 |
| Tetraodon nigroviridis | Query 732 DLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 767 | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 752 DRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 788 |
| Oryctolagus cuniculus | Query 732 DLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 767 | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 24 DRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 60 |
| Pongo pygmaeus | Query 732 DLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 767 | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 795 |
| Xenopus tropicalis | Query 732 DLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 767 | ||+ +|||||+ ++|+ +||++#||| |||+|||+ Sbjct 759 DRREFAKFEKEKMNAKWDTGENPIY#KSAVTTVVNPKY 795 |
| Pacifastacus leniusculus | Query 727 LTHLTDLREYRRFEKE-KLKSQWNNDNPLF#KSATTTVMNPKFAES 770 || | | ||| +|||| || | +|||+#||| || || ||+| Sbjct 756 LTTLHDRREYAKFEKERKLPSGKRAENPLY#KSAKTTFQNPAFAQS 800 |
| Anopheles gambiae str. PEST | Query 727 LTHLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 768 || + | ||+ |||||++ ++|+ +||++#| |||| || +| Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYA 835 |
| Anopheles gambiae | Query 727 LTHLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 768 || + | ||+ |||||++ ++|+ +||++#| |||| || +| Sbjct 793 LTSIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYA 835 |
| Aedes aegypti | Query 727 LTHLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 768 || + | ||+ |||||++ ++|+ +||++#| |||| || +| Sbjct 791 LTTIHDRREFARFEKERMMAKWDTGENPIY#KQATTTFKNPTYA 833 |
| Drosophila melanogaster | Query 727 LTHLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 768 || + | ||+ |||||++ ++|+ +||++#| ||+| || +| Sbjct 802 LTTIHDRREFARFEKERMNAKWDTGENPIY#KQATSTFKNPMYA 844 |
| Apis mellifera | Query 727 LTHLTDLREYRRFEKEK-LKSQWNN-DNPLF#KSATTTVMNPKF 767 || + | ||+++||+|+ | ++|| ||||+#| ||+| || | Sbjct 771 LTMIRDNREFKKFERERILANKWNRRDNPLY#KEATSTFKNPTF 813 |
| Caenorhabditis briggsae | Query 727 LTHLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 768 || | | || +| |+| ++|+ |+||++#| |||| || +| Sbjct 764 LTVLHDRAEYAKFNNERLMAKWDTNENPIY#KQATTTFKNPVYA 806 |
| Ostrinia furnacalis | Query 727 LTHLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 768 +| + | ||+ |||||++ ++|+ +||++#| ||+| || +| Sbjct 787 VTTIHDRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYA 829 |
| Strongylocentrotus purpuratus | Query 728 THLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAE 769 |++ | ||| ++| + |+||+ +|||++#||+||| || + + Sbjct 760 TYVQDKREYAQWENDCKKAQWDQSDNPIY#KSSTTTFKNPTYGK 802 |
| Tribolium castaneum | Query 728 THLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 767 | + | +|| +|| |+ | ||+ |||||+#+ ||+| || + Sbjct 711 TTVHDRQEYAKFENERQKMQWHRNDNPLY#RQATSTFKNPVY 751 |
| Caenorhabditis elegans | Query 727 LTHLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 768 || | | || | |+| ++|+ |+||++#| |||| || +| Sbjct 763 LTVLHDRSEYATFNNERLMAKWDTNENPIY#KQATTTFKNPVYA 805 |
| Pseudoplusia includens | Query 732 DLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFA 768 | ||+ |||||++ ++|+ +||++#| ||+| || +| Sbjct 798 DRREFARFEKERMMAKWDTGENPIY#KQATSTFKNPTYA 835 |
| Crassostrea gigas | Query 728 THLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAE 769 | ++| || ||||| + ++|+ +||++#| ||+| +|| + + Sbjct 757 TTISDKRELARFEKEAMNARWDTGENPIY#KQATSTFVNPTYRQ 799 |
| Biomphalaria glabrata | Query 727 LTHLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKF 767 || + | ||+ +||||+ ++|+ +||++#| ||+| || + Sbjct 744 LTFIHDTREFAKFEKERQNAKWDTGENPIY#KQATSTFKNPTY 785 |
| Acropora millepora | Query 730 LTDLREYRRFEKEKLKSQWNND-NPLF#KSATTTVMNPKFA 768 + | ||++||+|++ |+| + |||+#++| || || +| Sbjct 749 MVDRIEYQKFERERMHSKWTREKNPLY#QAAKTTFENPTYA 788 |
| Podocoryne carnea | Query 727 LTHLTDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKF 767 | + | ||+ +|||++ +|++ +||++#| ||+| || + Sbjct 737 LATIQDRREFAKFEKDRQNPKWDSGENPIY#KKATSTFQNPMY 778 |
| Lytechinus variegatus | Query 727 LTHLTDLREYRRFEKEKLKSQW-NNDNPLF#KSATTTVMNPKF 767 ||++ | ||++ ||||+ + | +||++#| +|+ || + Sbjct 762 LTYIHDRREFQNFEKERANATWEGGENPIY#KPSTSVFKNPTY 803 |
| Canis lupus familiaris | Query 727 LTHLTDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 764 | + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 778 |
| mice, Peptide Partial, 680 aa | Query 727 LTHLTDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 764 | + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 640 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 678 |
| rats, Peptide Partial, 723 aa | Query 727 LTHLTDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 764 | + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 679 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 717 |
| Equus caballus | Query 727 LTHLTDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMN 764 | + | +|+ +||+|+ +++|+ +|||+#| ||+| | Sbjct 740 LITIHDRKEFAKFEEERARAKWDTANNPLY#KEATSTFTN 778 |
| Schistosoma japonicum | Query 730 LTDLREYRRFEKEKLKSQWNN-DNPLF#KSATTTVMNPKFAES 770 + | || |+++ +| +||+|#+| || |+|| | |+ Sbjct 245 IDDRRELANFKQQGENMRWEMAENPIF#ESPTTNVLNPTFEEN 286 |
| Ophlitaspongia tenuis | Query 730 LTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAE 769 | |+ | |+||+| +++ |+|||+#+||| || + + Sbjct 798 LWDVVEVRKFEREIKNAKYTKNENPLY#RSATKDYQNPLYGK 838 |
| Geodia cydonium | Query 727 LTHLTDLREYRRFEKEKLKSQWN-NDNPLF#KSATTTVMNPKFAE 769 | + | | ++|| | | ++ |+|||+# |||| || + + Sbjct 835 LLMVLDYTELKKFENELAKVKYTKNENPLY#HSATTEHKNPIYGQ 878 |
* References
[PubMed ID: 19299700] Sorgi CA, Secatto A, Fontanari C, Turato WM, Belanger C, de Medeiros AI, Kashima S, Marleau S, Covas DT, Bozza PT, Faccioli LH, Histoplasma capsulatum cell wall {beta}-glucan induces lipid body formation through CD18, TLR2, and dectin-1 receptors: correlation with leukotriene B4 generation and role in HIV-1 infection. J Immunol. 2009 Apr 1;182(7):4025-35.
[PubMed ID: 19273622] ... Jakus Z, Simon E, Frommhold D, Sperandio M, Mocsai A, Critical role of phospholipase Cgamma2 in integrin and Fc receptor-mediated neutrophil functions and the effector phase of autoimmune arthritis. J Exp Med. 2009 Mar 16;206(3):577-93. Epub 2009 Mar 9.
[PubMed ID: 19234461] ... Moser M, Bauer M, Schmid S, Ruppert R, Schmidt S, Sixt M, Wang HV, Sperandio M, Fassler R, Kindlin-3 is required for beta2 integrin-mediated leukocyte adhesion to endothelial cells. Nat Med. 2009 Mar;15(3):300-5. Epub 2009 Feb 22.
[PubMed ID: 19201847] ... Buhtoiarov IN, Rakhmilevich AL, Lanier LL, Ranheim EA, Sondel PM, Naive mouse macrophages become activated following recognition of L5178Y lymphoma cells via concurrent ligation of CD40, NKG2D, and CD18 molecules. J Immunol. 2009 Feb 15;182(4):1940-53.
[PubMed ID: 19066088] ... Titova E, Kevil CG, Ostrowski RP, Rojas H, Liu S, Zhang JH, Tang J, Deficiency of CD18 gene reduces brain edema in experimental intracerebral hemorrhage in mice. Acta Neurochir Suppl. 2008;105:85-7.
[PubMed ID: 1431091] ... Morse HC 3rd, Genetic nomenclature for loci controlling surface antigens of mouse hemopoietic cells. J Immunol. 1992 Nov 15;149(10):3129-34.
[PubMed ID: 1783381] ... Threadgill DS, Womack JE, Mapping HSA 3 loci in cattle: additional support for the ancestral synteny of HSA 3 and 21. Genomics. 1991 Dec;11(4):1143-8.
[PubMed ID: 1837533] ... MacDonald G, Chu ML, Cox DR, Fine structure physical mapping of the region of mouse chromosome 10 homologous to human chromosome 21. Genomics. 1991 Oct;11(2):317-23.
[PubMed ID: 1671669] ... Kofler R, Mouse macrophage beta subunit (CD11b) cDNA for the CR3 complement receptor/Mac-1 antigen. Immunogenetics. 1991;33(1):77-8.
[PubMed ID: 1969385] ... Zeger DL, Osman N, Hennings M, McKenzie IF, Sears DW, Hogarth PM, Mouse macrophage beta subunit (CD11b) cDNA for the CR3 complement receptor/Mac-1 antigen. Immunogenetics. 1990;31(3):191-7.