XSB0998 : growth associated protein 43 isoform 2 [Homo sapiens]
[ CaMP Format ]
This entry is computationally expanded from SB0067
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | Neuromodulin; Axonal membrane protein GAP-43; Growth-associated protein 43; PP46; Neural phosphoprotein B-50; growth associated protein 43 isoform 2; neuron growth-associated protein 43; nerve growth-related peptide GAP43; protein F1; calmodulin-binding protein P-57 |
| Gene Names | GAP43; growth associated protein 43 |
| Gene Locus | 3q13.1-q13.2; chromosome 3 |
| GO Function | Not available |
* Information From OMIM
Function: Chen et al. (2003) used cDNA microarrays to measure the expression of multiple genes in primary cultured rat hippocampal cells exposed to 1 mmol/L desipramine, 2 mmol/L tranylcypromine, 1 mmol/L lithium, and 10 mmol/L haloperidol. In situ hybridization revealed that desipramine increased GAP43 gene expression in dentate gyrus but not other brain regions. Northern and immunoblotting analyses showed that GAP43 mRNA and protein levels were increased in response to both desipramine and tranylcypromine but not in response to lithium or haloperidol. Because GAP43 regulates the growth of axons and modulates the formation of new connections, Chen et al. (2003) suggested that desipramine may have an effect on neuronal plasticity in the CNS.
* Structure Information
1. Primary Information
Length: 238 aa
Average Mass: 24.803 kDa
Monoisotopic Mass: 24.788 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Neuromodulin_N 1. | 1 | 31 | 65.8 | 1.6e-16 |
| --- cleavage 4 (inside Neuromodulin_N 1..31) --- | ||||
| IQ 1. | 32 | 52 | 32.4 | 1.9e-06 |
| Neuromodulin 1. | 67 | 238 | 353.9 | 3e-103 |
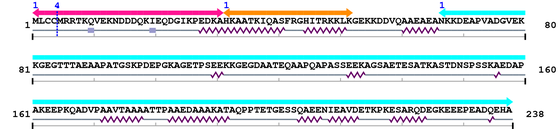
3. Sequence Information
Fasta Sequence: XSB0998.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] MLCC4-MRRTKQVEKN
Cys4  Met
Met
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | Met1 | Leu2 | Cys3 | Cys4 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Met5 | Arg6 | Arg7 | Thr8 | Lys9 | Gln10 | Val11 | Glu12 | Lys13 | Asn14 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 17577668] Bolognani F, Tanner DC, Nixon S, Okano HJ, Okano H, Perrone-Bizzozero NI, Coordinated expression of HuD and GAP-43 in hippocampal dentate granule cells during developmental and adult plasticity. Neurochem Res. 2007 Dec;32(12):2142-51. Epub 2007 Jun 19.