XSB2230 : glucagon preproprotein [Homo sapiens]
[ CaMP Format ]
This entry is computationally expanded from SB0054
* Basic Information
| Organism | Homo sapiens (human) |
| Protein Names | Glucagon; Glicentin; Glicentin-related polypeptide; GRPP; Oxyntomodulin; OXY; OXM; Glucagon-like peptide 1; GLP-1; Glucagon-like peptide 1(7-37); GLP-1(7-37); Glucagon-like peptide 1(7-36); GLP-1(7-36); Glucagon-like peptide 2; GLP-2; glucagon preproprotein; RecName: Full=Glucagon; Contains:RecName: Full=Glicentin; Contains:RecName: Full=Glicentin-related polypeptide; Short=GRPP; Contains:RecName: Full=Oxyntomodulin; Short=OXY; Short=OXM; Contains:RecName: Full=Glucagon; Contains:RecName: Full=Glucagon-like peptide 1; Short=GLP-1; 7-37; 7-36 |
| Gene Names | GCG; glucagon |
| Gene Locus | 2q36-q37; chromosome 2 |
| GO Function | Not available |
* Information From OMIM
Description: Glucagon is a 29-amino acid pancreatic hormone that counteracts the glucose-lowering action of insulin by stimulating glycogenolysis and gluconeogenesis. Human, rabbit, rat, pig, and cow glucagons are identical. Glucagon is a member of a multigene family that includes secretin (OMIM:182099), vasoactive intestinal peptide (VIP; OMIM:192320), gastric inhibitory peptide (GIP; OMIM:137240), glicentin, and others.
Function: GLP1, also known as 7-37 for the codons of the preproglucagon molecule which encode it, renders pancreatic beta-cells 'glucose-competent' and may be useful in the treatment of noninsulin-dependent diabetes mellitus (Holz et al., 1993).
Function: GLP1 is a potent insulin secretagogue. Wang et al. (1995) presented evidence that it plays a major role in the enteroinsular axis, accounting, for example, for the finding that plasma insulin levels accompanying oral intake of glucose are greater than those observed when glucose is given intravenously. It is the so-called gluco-incretin. Wang et al. (1995) used an inhibitor of GLP1, called exendin(9-39), which is a fragment of a peptide found in venom of gila monsters that binds tightly to the GLP1 receptor (GLP1R; OMIM:138032) without agonistic activity. Although GLP1 and its specific receptors are present in the hypothalamus, no physiologic role for central GLP1 had been established. Turton et al. (1996) found that intracerebroventricular GLP1 powerfully inhibited feeding in fasted rats. Injection of the specific antagonist, exendin, blocked the inhibitory effect of GLP1 on food intake. Exendin alone had no influence on fast-induced feeding but more than doubled food intake in satiated rats, and augmented the feeding response to the appetite stimulus, neuropeptide Y (OMIM:162640). Induction of FOS (OMIM:164810) is a marker of neuronal activation. Following intracerebroventricular GLP1 injection, FOS appeared exclusively in the paraventricular nucleus of the hypothalamus and central nucleus of the amygdala, and this was inhibited by prior administration of exendin. Both of these regions of the brain are of primary importance in the regulation of feeding. The findings suggested that central GLP1 is a physiologic mediator of satiety.
Function: Drucker (1999) reviewed the physiologic relevance of GLP2. GLP2 is secreted in a nutrient-dependent manner from enteroendocrine cells throughout the gastrointestinal tract and is trophic to the intestinal epithelial mucosa. It acts via stimulation of crypt cell proliferation and inhibition of cell death. GLP2 also stimulates intestinal glucose transport, decreases mucosal permeability, and has shown therapeutic efficacy in experimental models of short bowel syndrome and both small and large bowel inflammation.
Function: Drucker (2001) noted that both GLP1 and GLP2 are secreted from gut endocrine cells and promote nutrient absorption through distinct mechanisms. GLP2 regulates gastric motility, gastric acid secretion, intestinal hexose transport, and increases the barrier function of the gut epithelium. GLP2 significantly enhances the surface area of the mucosal epithelium via stimulation of crypt cell proliferation and inhibition of apoptosis in the enterocyte and crypt compartments. The cytoprotective and reparative effects of GLP2 are evident in rodent models of experimental intestinal injury. GLP2 reduces mortality and decreases mucosal injury, cytokine expression, and bacterial septicemia in the setting of small and large bowel inflammation. GLP2 also enhances nutrient absorption and gut adaptation in rodents or humans with short bowel syndrome. The actions of GLP2 are transduced by the GLP2 receptor (OMIM:603659), a G protein-coupled receptor expressed in gut endocrine cells of the stomach, small bowel, and colon. Activation of GLP2 receptor signaling in heterologous cells promotes resistance to apoptotic injury in vitro. The authors concluded that the cytoprotective, reparative, and energy-retentive properties of GLP2 suggest that GLP2 may potentially be useful for the treatment of human disorders characterized by injury and/or dysfunction of the intestinal mucosal epithelium.
Function: GLP2 is metabolized extensively by dipeptidyl peptidase IV (DPP4; OMIM:102720) in rats. To elucidate its fate in humans, Hartmann et al. (2000) investigated GLP2 metabolism in healthy volunteers after (1) a 500-calorie mixed meal; (2) intravenous infusion of synthetic human GLP2; (3) a subcutaneous bolus injection; and (4) in vitro incubation in plasma and blood. GLP2 concentrations were determined by N-terminal RIA measuring only intact GLP2, side-viewing RIA measuring intact and degraded forms (e.g., GLP2-(3-33) arising from DPP4 degradation), and high performance liquid chromatography (HPLC). Meal ingestion elevated plasma GLP2 (intact, 16 +/- 3 to 73 +/- 10 pmol/L at 90 minutes), and HPLC revealed 2 immunoreactive components, intact GLP2 and GLP2-(3-33). The elimination t-1/2 values were 7.2 +/- 2 min (intact GLP2) and 27.4 +/- 5.4 min (GLP2-(3-33)), and MCRs were 6.8 +/- 0.6 and 1.9 +/- 0.3 mL/kg.min, respectively. Subcutaneous injection increased intact GLP2 to maximally 1,493 +/- 250 pmol/L at 45 minutes, whereas total GLP2 increased to 2,793 +/- 477 pmol/L at 90 minutes. Hartmann et al. (2000) concluded that GLP2 is extensively degraded to GLP2-(3-33) in humans, presumably by DPP4. Nevertheless, 69% remained intact 1 hour after GLP2 injection, supporting the possibility of subcutaneous use in patients with intestinal insufficiency.
Function: Pancreatic beta cells share several molecular mechanisms with pancreatic endocrine cells in terms of development, therefore they may possess a potential for insulin expression. To test this idea, Suzuki et al. (2003) sought to induce insulin-production in intestinal epithelial progenitors by using glucagon-like peptide-1. They found that GLP1-(1-37) induces insulin production in developing and, to a lesser extent, adult intestinal epithelial cells in vitro and in vivo, a process mediated by upregulation of the Notch-related gene encoding neurogenin-3 (NGN3; OMIM:604882) and its downstream targets, which are involved in pancreatic endocrine differentiation. These cells became responsive to glucose challenge in vitro and reverse insulin-dependent diabetes after implantation into diabetic mice. The findings suggested that efficient induction of insulin production in intestinal epithelial cells by GLP1-(1-37) could represent a new therapeutic approach for diabetes mellitus. The experiments were performed in the cell and organ culture.
Function: Hirasawa et al. (2005) found colocalization of GPR120 (OMIM:609044) and GLP1 in human colonic intraepithelial neuroendocrine cells. Using several techniques, including RNA interference, they showed that mouse Gpr120 mediated free fatty acid-induced secretion of Glp1 in a mouse intestinal endocrine cell line.
Function: Dyachok et al. (2006) introduced a new ratiometric evanescent-wave-microscopy approach to measure cAMP concentration beneath the plasma membrane and showed that insulin-secreting beta cells respond to glucagon and GLP1 with marked cAMP oscillations. Simultaneous measurements of intracellular calcium concentration revealed that the 2 messengers are interlinked and reinforce each other. Moreover, cAMP oscillations are capable of inducing rapid on-off calcium responses, but only sustained elevation of cAMP concentration induces nuclear translocation of the catalytic subunit of the cAMP-dependent protein kinase. Dyachok et al. (2006) concluded that their results established a new signaling mode for cAMP and indicated that temporal encoding of cAMP signals might constitute a basis for differential regulation of downstream cellular targets.
Function: Jang et al. (2007) examined human duodenal biopsy specimens and found expression of the sweet taste receptors T1R2 (OMIM:606226) and T1R3 (OMIM:605865), alpha-gustducin (GNAT3; OMIM:139395), and other taste transduction elements in the enteroendocrine L cells. Mouse intestinal cells also expressed alpha-gustducin, and ingestion of glucose by Gnat3-null mice revealed deficiencies in secretion of GLP1 and the regulation of plasma insulin (OMIM:176730) and glucose. Isolated small bowel and intestinal villi from Gnat3-null mice showed markedly defective GLP1 secretion in response to glucose. In human NCI-H716 L cells, GLP1 release was promoted by sugars and the artificial sweetener sucralose and blocked by the sweet-receptor antagonist lactisole or siRNA for alpha-gustducin. Jang et al. (2007) concluded that L cells of the gut 'taste' glucose through the same mechanism used by taste cells of the tongue.
Function: Margolskee et al. (2007) demonstrated that dietary sugar and artificial sweeteners increased sodium-dependent glucose transporter SGLT1 (SLC5A1; OMIM:182380) mRNA and protein expression and increased glucose absorptive capacity in wildtype mice but not in knockout mice lacking T1R3 or alpha-gustducin. In mouse GLUTag enteroendocrine cells, sucralose increased the release of GLP1 and GIP (OMIM:137240), gut hormones implicated in SGLT1 upregulation, and increased intracellular calcium; inhibition of the T1R2-T1R3 sweet taste receptor by gurmarin blocked the sucralose-stimulated release of GLP1 and GIP and the sucralose-dependent mobilization of calcium in GLUTag cells.
* Structure Information
1. Primary Information
Length: 180 aa
Average Mass: 20.909 kDa
Monoisotopic Mass: 20.896 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Hormone_2 1. | 53 | 80 | 66.4 | 1.1e-16 |
| --- cleavage 74 (inside Hormone_2 53..80) --- | ||||
| --- cleavage 69 (inside Hormone_2 53..80) --- | ||||
| --- cleavage 77 (inside Hormone_2 53..80) --- | ||||
| --- cleavage 67 (inside Hormone_2 53..80) --- | ||||
| --- cleavage 79 (inside Hormone_2 53..80) --- | ||||
| Hormone_2 2. | 98 | 125 | 52.6 | 1.5e-12 |
| Hormone_2 3. | 146 | 173 | 45.7 | 1.9e-10 |
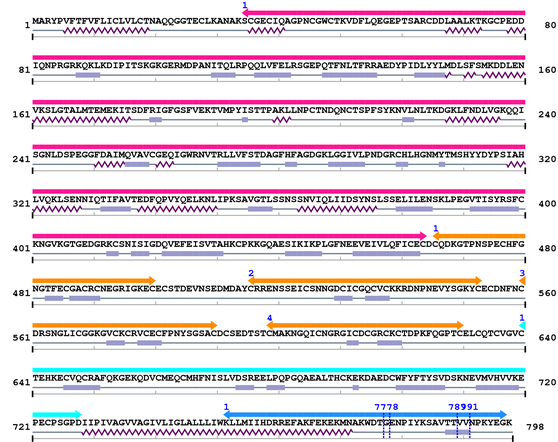
3. Sequence Information
Fasta Sequence: XSB2230.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
* Cleavage Information
5 [sites]
Cleavage sites (±10aa)
[Site 1] YLDSRRAQDF74-VQWLMNTKRN
Phe74  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Tyr65 | Leu66 | Asp67 | Ser68 | Arg69 | Arg70 | Ala71 | Gln72 | Asp73 | Phe74 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val75 | Gln76 | Trp77 | Leu78 | Met79 | Asn80 | Thr81 | Lys82 | Arg83 | Asn84 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DQMNEDKRHSQGTFTSDYSKYLDSRRAQDFVQWLMNTKRNRNNIAKRHDEFERHAEGTFT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 129.00 | 10 | glucagon, isoform CRA_b |
| 2 | N/A | 79.70 | 44 | 0 |
| 3 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | 64.70 | 1 | AAB31092.1 |
| 4 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | 61.60 | 1 | glucagon=proglucagon-derived 3.5 kda peptide |
| 5 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | 60.80 | 1 | glucagon II |
| 6 | Contains: Glucagon; Glucagon-like peptide | 60.10 | 4 | GLUC_AMICA Glucagon precursor |
| 7 | Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | 60.10 | 1 | glucagon |
| 8 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | 58.50 | 1 | glucagon G-29 |
| 9 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | 57.80 | 1 | GLUC1_ORENI Glucagon-1 (Glucagon I) gb |
| 10 | Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | 56.20 | 1 | GLUC_LAMFL Glucagon gb |
| 11 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | 53.50 | 1 | glucagon |
| 12 | deshis1, Desphe6, Glu9 | 53.50 | 1 | A Chain A, Nmr Solution Structure Of The Glucagon |
| 13 | Monodelphis domestica | 48.90 | 3 | PREDICTED: similar to Gastric inhibitory polypepti |
| 14 | Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | 48.90 | 1 | GLUC1_PETMA Glucagon-1 precursor |
| 15 | Xenopus laevis | 47.00 | 5 | proglucagon II |
| 16 | Hoplobatrachus rugulosus | 46.60 | 1 | AF324209_1 proglucagon |
| 17 | Gallus gallus | 46.20 | 4 | glucagon isoform 1 |
| 18 | Meleagris gallopavo | 46.20 | 2 | preproglucagon B |
| 19 | Bufo marinus | 44.70 | 1 | proglucagon |
| 20 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | 44.70 | 1 | GLUC_HELSU Glucagon precursor |
| 21 | Capra hircus | 43.90 | 1 | glucagon |
| 22 | Oncorhynchus tshawytscha | 43.50 | 1 | S78474_1 proglucagon |
| 23 | Ictalurus punctatus | 43.10 | 3 | proglucagon I |
| 24 | Rattus norvegicus | 42.70 | 5 | Glucose-dependent insulinotropic peptide |
| 25 | Canis familiaris | 42.70 | 2 | PREDICTED: similar to Gastric inhibitory polypepti |
| 26 | Xenopus tropicalis | 42.40 | 2 | glucagon |
| 27 | Pan troglodytes | 42.40 | 2 | PREDICTED: fibroblast activation protein, alpha su |
| 28 | Danio rerio | 42.00 | 6 | hypothetical protein LOC494052 |
| 29 | Bos taurus | 42.00 | 5 | vasoactive intestinal peptide |
| 30 | Macaca mulatta | 41.60 | 3 | PREDICTED: vasoactive intestinal peptide |
| 31 | Heloderma suspectum | 41.60 | 2 | proglucagon |
| 32 | Squalus acanthias | 41.60 | 1 | proglucagon |
| 33 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | 41.20 | 1 | glucagon-like peptide, GLP |
| 34 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | 41.20 | 1 | GLUC2_LOPAM Glucagon-2 precursor (Glucagon II) |
| 35 | Sebastes caurinus | 40.80 | 4 | proglucagon I |
| 36 | Tetraodon nigroviridis | 40.00 | 3 | unnamed protein product |
| 37 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | 40.00 | 2 | GLUC_CARAU Glucagon precursor |
| 38 | Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC1_ONCMY Glucagon-1 precursor (Glucagon I) |
| 39 | Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC2_ONCMY Glucagon-2 precursor (Glucagon II) |
| 40 | Heloderma horridum | 40.00 | 1 | EXE3_HELHO Exendin-3 precursor emb |
| 41 | Mus musculus | 39.70 | 6 | Gastric inhibitory polypeptide |
| 42 | Protopterus dolloi | 38.90 | 1 | proglucagon |
| 43 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | 38.90 | 1 | glucagon-like peptide-2, GLP-2=proglucagon-derived |
| 44 | Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | 38.50 | 1 | VIP_MOUSE VIP peptides precursor |
| 45 | Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | 38.10 | 1 | GLUC2_PETMA Glucagon-2 precursor |
| 46 | Ambloplites rupestris | 37.70 | 1 | proglucagon |
| 47 | Arvicanthis ansorgei | 37.70 | 1 | vasoactive intestinal polypeptide |
| 48 | synthetic construct | 37.00 | 1 | Homo sapiens glucagon |
| 49 | Phodopus sungorus | 36.60 | 1 | glucagon precursor |
| 50 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | 36.60 | 1 | GLUC1_LOPAM Glucagon-1 precursor (Glucagon I) |
| 51 | Sus scrofa | 36.20 | 1 | glucagon |
| 52 | Neoceratodus forsteri | 36.20 | 1 | proglucagon |
| 53 | Canis lupus familiaris | 35.80 | 1 | glucagon |
| 54 | Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | 35.80 | 1 | GLUC_LEPSP Glucagon precursor |
| 55 | Hydrolagus colliei | 35.40 | 1 | proglucagon |
| 56 | Heloderma suspectum, venom, Peptide, 39 aa | 35.40 | 1 | A Chain A, Solution Structure Of Exendin-4 In 30-V |
| 57 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | 34.70 | 1 | GLUC2_ORENI Glucagon-2 (Glucagon II) gb |
| 58 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | 34.70 | 1 | glucagon-like peptide-1, GLP-1=proglucagon-derived |
| 59 | Agkistrodon piscivorus | 33.50 | 1 | proglucagon |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 |
| N/A | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIA 89 ||||||||||||||||||||||#||||||||||+|||| Sbjct 1 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIA 37 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRN 84 ||+||||||||||+|+|||+||#|||||+|||| Sbjct 1 HSEGTFTSDYSKYMDNRRAKDF#VQWLMSTKRN 32 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 |||||||||||||||+||||||#+||||+| Sbjct 1 HSQGTFTSDYSKYLDNRRAQDF#IQWLMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRN 84 ||+|||++||||||++||||||#|||| |+||| Sbjct 1 HSEGTFSNDYSKYLETRRAQDF#VQWLKNSKRN 32 |
| Contains: Glucagon; Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 |||||||+|||||+|+||||||#|||||+| Sbjct 1 HSQGTFTNDYSKYMDTRRAQDF#VQWLMST 29 |
| Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 |||||||+|||||+|+||||||#|||||+| Sbjct 1 HSQGTFTNDYSKYMDTRRAQDF#VQWLMST 29 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 ||+||||||||||+|+|||+||#|||||+| Sbjct 1 HSEGTFTSDYSKYMDNRRAKDF#VQWLMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRN 84 ||+|||++||||||+ |+||||#|+|||| ||+ Sbjct 1 HSEGTFSNDYSKYLEDRKAQDF#VRWLMNNKRS 32 |
| Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 ||||+|||||||||||++|+||#| ||||| Sbjct 1 HSQGSFTSDYSKYLDSKQAKDF#VIWLMNT 29 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMN 80 ||+|||||||||||++++|+||#|+|||| Sbjct 1 HSEGTFTSDYSKYLENKQAKDF#VRWLMN 28 |
| deshis1, Desphe6, Glu9 | Query 54 SQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 |||| ||+|||||||||||||#||||||| Sbjct 1 SQGT-TSEYSKYLDSRRAQDF#VQWLMNT 27 |
| Monodelphis domestica | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGT 102 +|+++||| |||| +| |||#| ||++ | +| + ||+ || |+|| Sbjct 51 QRYAEGTFISDYSITMDKIMQQDF#VNWLLSQKGKKN--SWRHNITERKADGT 100 |
| Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ||||+|||||||||||++++|+||#|+|||| || + + + |||+|||| Sbjct 41 KRHSEGTFTSDYSKYLENKQAKDF#VRWLMNAKRGGSELQR------RHADGTFT 88 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNR---NNIAKRHDEFERHAE 100 +||+ ||||+| + |||++ |+||#| || + ++| + + | +|||| Sbjct 80 RRHADGTFTNDMTSYLDAKAARDF#VSWLARSDKSRRDGGDHLAENSEDKRHAE 132 |
| Xenopus laevis | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 +|+ | ||||||||||||||||||||||||#|||||||||+ +++|+ ++||||||||| Sbjct 45 EQLKEVKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRS-GGLSRRNADYERHAEGTFT 103 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 | +||++||+|+| ++||+ + + |#++||+ | + + |||||||| Sbjct 138 EIERHAEGTYTNDVTEYLEEKATKAF#IEWLIKGK-------PKKIRYSRHAEGTFT 186 Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRH 92 ||++||||+| + ||+ + |++|#| ||+| + | ++ + | Sbjct 179 RHAEGTFTNDMTNYLEEKAAKEF#VGWLINGRPKRKDLLEEH 219 |
| Hoplobatrachus rugulosus | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 | || ||||||||||||||||||||||||#||||||+||+ |+||+ +||||||||+| Sbjct 45 DHHNEVKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNSKRS-GGISKRNVQFERHAEGTYT 103 Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFE-RHAEGTFT 104 ||++|||||| + ||+ + |++|#| ||+ + || +| + |||+|+|| Sbjct 134 RHAEGTFTSDMTSYLEEKAAKEF#VDWLIKGRPKRNFPDISAEEMDRRHADGSFT 187 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 +||++||+|+| +++|+ + |++|#+ ||+ | + ++ |||||||| Sbjct 95 ERHAEGTYTNDVTQFLEEKAAKEF#IDWLIKGKPKKQRLS-------RHAEGTFT 141 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 |+||+ |+||||++| || + ||+|#+ |++|| Sbjct 178 DRRHADGSFTSDFNKALDIKAAQEF#LDWIINT 209 |
| Gallus gallus | Query 46 QMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRN------------------RNN 87 |+|| ||||||||||||||||||||||||#|||||+|||| | Sbjct 48 QLNEVKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMSTKRNGQQGQEDKENDKFPDQLSSNA 107 Query 88 IAKRHDEFERHAEGTFT 104 |+||| |||||||||+| Sbjct 108 ISKRHSEFERHAEGTYT 124 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAK---RHDEFERHAEGTFT 104 +| +||++||+||| + ||+ + |++|#+ ||+| + |+ | + |||+|||| Sbjct 113 SEFERHAEGTYTSDITSYLEGQAAKEF#IAWLVNGRGRRDFPEKALMAEEMGRRHADGTFT 172 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNI 88 +||+ |||||| +| || |++|#++||+||| + ++ Sbjct 164 RRHADGTFTSDINKILDDMAAKEF#LKWLINTKVTQRDL 201 |
| Meleagris gallopavo | Query 46 QMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRN------------------RNN 87 |+|| ||||||||||||||||||||||||#|||||+|||| | Sbjct 48 QLNEVKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMSTKRNGQQGQEDKENDEFPDQLSSNA 107 Query 88 IAKRHDEFERHAEGTFT 104 |+||| |||||||||+| Sbjct 108 ISKRHSEFERHAEGTYT 124 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAK---RHDEFERHAEGTFT 104 +| +||++||+||| + ||+ + |++|#+ ||+| + |+ | + |||+|||| Sbjct 113 SEFERHAEGTYTSDITSYLEGQAAKEF#IAWLVNGRGRRDFPEKALMAEEMGRRHADGTFT 172 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNI 88 +||+ |||||| +| || |++|#++||+||| + ++ Sbjct 164 RRHADGTFTSDINKILDDMAAKEF#LKWLINTKVTQRDL 201 |
| Bufo marinus | Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLM--NTKRNRNNIAKRHDEFERHAEGTFT 104 ||++|||||| + +|+ + |++|#| ||+ ||| ++++ | |||+|+|| Sbjct 62 RHAEGTFTSDMTSFLEEKAAKEF#VDWLIKGRPKRNFSDVSAADDMDRRHADGSFT 116 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 +||++||+|+| +++|+ + |++|#+ ||+ | |+ |||||||| Sbjct 23 ERHAEGTYTNDVTQFLEEKAAKEF#IDWLL------KGIPKK-QRLSRHAEGTFT 69 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 |+||+ |+||||++| || + ||+|#+ |++|| Sbjct 107 DRRHADGSFTSDFNKALDIKAAQEF#LDWIINT 138 Query 73 DF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ||# |||||+||+ +++|+ +||||||||+| Sbjct 1 DF#AQWLMNSKRS-GGMSRRNVQFERHAEGTYT 31 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | Query 46 QMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKR------------------NRNN 87 |+|| |||||||||||||||||+||||||#||||||||| + | Sbjct 46 QLNEVKRHSQGTFTSDYSKYLDTRRAQDF#VQWLMNTKRSGQQGVEEREKENLLDQLSSNG 105 Query 88 IAKRHDEFERHAEGTFT 104 +|+ | |+||||+| +| Sbjct 106 LARHHAEYERHADGRYT 122 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAK---RHDEFERHAEGTFT 104 | +||+ | +||| | ||+ + |++|#+ ||+| + |+ + + | |||+|||| Sbjct 112 EYERHADGRYTSDISSYLEGQAAKEF#IAWLVNGRGRRDFLEEAGTADDIGRRHADGTFT 170 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNI 88 +||+ |||||||++ || |+|#++||+| | + ++ Sbjct 162 RRHADGTFTSDYNQLLDDIATQEF#LKWLINQKVTQRDL 199 |
| Capra hircus | Query 86 NNIAKRHDEFERHAEGTFT 104 ||||||||||||||||||| Query 86 NNIAKRHDEFERHAEGTFT 104 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNR 85 +| +||++|||||| | ||+ + |++|#+ ||+ + | Sbjct 8 DEFERHAEGTFTSDVSSYLEGQAAKEF#IAWLVKGRGRR 45 |
| Oncorhynchus tshawytscha | Query 54 SQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 | |||++||||| + | ||||#||||||+||+ +|||+||+| Sbjct 2 SXGTFSNDYSKYQEERMAQDF#VQWLMNSKRS-------GAPSKRHADGTYT 45 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNR 85 |||+ ||+||| | || + |+||#| || + + | Sbjct 37 KRHADGTYTSDVSTYLQDQAAKDF#VSWLKSGRARR 71 |
| Ictalurus punctatus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 +|||+|||++||||||++||||||#||||||+||+ |||+||+| Sbjct 49 RRHSEGTFSNDYSKYLETRRAQDF#VQWLMNSKRS-------DSPARRHADGTYT 95 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEF--ERHAEGTFT 104 +||+ ||+||| | || + |+||#+ || + + + + | + || +|+|| Sbjct 87 RRHADGTYTSDVSSYLQDQAAKDF#ITWLKSGQPKPDAVETRSVDTLRRRHVDGSFT 142 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRH 92 +|| |+|||| +| ||| |+++#+||+||+ + + ||| Sbjct 134 RRHVDGSFTSDVNKVLDSLAAKEY#LQWVMNSPSSAS--GKRH 173 |
| Rattus norvegicus | Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHA 99 |+++||| |||| +| | |||#| ||+ | +|+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDF#VNWLLAQKGKKND--WKHNLTQREA 88 |
| Canis familiaris | Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHA 99 |+++||| |||| +| | |||#| ||+ | +|+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDF#VNWLLAQKGKKND--WKHNITQREA 88 |
| Xenopus tropicalis | Query 46 QMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 |+ | ||||||||||||||||||||||||#+||||||||+ +++|+ ++||||||||| Sbjct 46 QLKEVKRHSQGTFTSDYSKYLDSRRAQDF#IQWLMNTKRS-GGLSRRNADYERHAEGTFT 103 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 +||++|||||| +++|| + |++|#+ ||+| ++ |++|+ + |||||||+| Sbjct 95 ERHAEGTFTSDVTQHLDEKAAKEF#IDWLINGGPSKEIISRRNADVERHAEGTYT 148 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 +||++||+|+| ++||+ + |++|#++||+| | + + |||||||| Sbjct 140 ERHAEGTYTNDVTEYLEEKAAKEF#IEWLINGK-------PKKIRYSRHAEGTFT 186 Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFE---RHAEGTFT 104 ||++||||+| + ||+ + |++|#| ||+ | + | ++ | | |||+|+|| Sbjct 179 RHAEGTFTNDMTNYLEEKAAKEF#VGWLIK-GRPKRNFSEAHSVEEMDRRHADGSFT 233 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 |+||+ |+||+| +| || ||+|#+ |++||+ Sbjct 224 DRRHADGSFTNDINKVLDIIAAQEF#LDWVINTQ 256 |
| Pan troglodytes | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ||||||||||||||||||||||||||||||#|||||||||| +++| |||||||| Sbjct 967 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRN-SSVA------YRHAEGTFT 1019 Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNR 85 ||++|||||| | ||+ + |++|#+ ||+ + | Sbjct 1012 RHAEGTFTSDVSSYLEGQAAKEF#IAWLVKGRGRR 1045 |
| Danio rerio | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ||||+|||++||||||++||||||#|||||| ||| ++ +||||||+| Sbjct 48 KRHSEGTFSNDYSKYLETRRAQDF#VQWLMNAKRNGGSV-------KRHAEGTYT 94 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWL 78 |||++||+||| | || + || |#| || Sbjct 86 KRHAEGTYTSDVSSYLQDQAAQSF#VAWL 113 |
| Bos taurus | Query 47 MNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ++ + ||+ | ||||||+ | |+ +#++ |+ || |+|++ +||++ || Sbjct 75 VSRNVRHADGVFTSDYSRLLGQLSAKKY#LESLIG-KRVSNSISEDQGPIKRHSDAVFT 131 |
| Macaca mulatta | Query 47 MNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ++ + ||+ | ||||+|| | |+ +#++ || || +||++ +||++ || Sbjct 75 VSRNARHADGVFTSDFSKLLGQLSAKKY#LESLMG-KRVSSNISEDPVPVKRHSDAVFT 131 |
| Heloderma suspectum | Query 46 QMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKR------------------NRNN 87 |+|| |||||||||||||||||+||||||#||||||||| + | Sbjct 46 QLNEVKRHSQGTFTSDYSKYLDTRRAQDF#VQWLMNTKRSGQQGVEEREKENLLDQLSSNG 105 Query 88 IAKRHDEFERHAEGTFT 104 +|+ | |+||||+| +| Sbjct 106 LARHHAEYERHADGRYT 122 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNR 85 | +||+ | +||| | ||+ + |++|#+ ||+| + | Sbjct 112 EYERHADGRYTSDISSYLEGQAAKEF#IAWLVNGRGRR 148 |
| Squalus acanthias | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWL--MNTKRNRNNIAKRHDEFERHAEGTF 103 ++ |||++||+||| | +|+ |#| | | ++| +|+||| | || ||++ Sbjct 20 DKTKRHAEGTYTSDVDSLSDYFKAKRF#VDSLTSYNKRQNGRSISKRHVESTRHTEGSY 77 Query 67 DSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 |+|||+||#|||||+|||| |+ +||||||+| Sbjct 1 DNRRAKDF#VQWLMSTKRN-------GDKTKRHAEGTYT 31 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRN---------NIAKRHDEFERHA 99 | ||++|++ | | ||+++ |+||#+ ||+ + | | + ||| Sbjct 68 ESTRHTEGSYR-DISSYLEAKAAKDF#INWLIKGRGRREFPEESKEIVNEVIPEEMDRRHA 126 Query 100 EGTFT 104 +|||| Sbjct 127 DGTFT 131 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFE 96 |+||+ |||||+ + ||+ |++|#+ |++|+| | | | | Sbjct 122 DRRHADGTFTSEINIVLDTIAAKEF#LNWILNSK----TIQSRDSEIE 164 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRN 84 |+ ||||+| + |||++ |+||#| || + ++ Sbjct 1 HADGTFTNDMTSYLDAKAARDF#VSWLARSDKS 32 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ||||+|||++||||||++||||||#|||| |+||| | |||+||+| Sbjct 50 KRHSEGTFSNDYSKYLETRRAQDF#VQWLKNSKRN--------GLFRRHADGTYT 95 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWL 78 +||+ ||+||| | || + |+||#| || Sbjct 87 RRHADGTYTSDVSSYLQDQAAKDF#VSWL 114 |
| Sebastes caurinus | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 |+++ ||||+|||++|||+||+ |+||||#|+|||| ||+ || +|||+|||| Sbjct 42 DELSNMKRHSEGTFSNDYSRYLEERKAQDF#VRWLMNNKRS------GADE-KRHADGTFT 94 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDE--FERHAEGTFT 104 ++|||+ |||||| | || + +||#| | + + | + + | || +|+|| Sbjct 84 DEKRHADGTFTSDVSSYLKDQAIKDF#VNRLKSGQVRRESETEWRGEAFSRRHVDGSFT 141 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDE 94 +|| |+|||| +| ||| |+++#+ |+| +| + + ++ |+ Sbjct 133 RRHVDGSFTSDVNKVLDSMAAKEY#LLWVMTSKPSGESKKRQEDQ 176 |
| Tetraodon nigroviridis | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ||||+|||++||||||++||||||#|||| |+||| + | |||+||+| Sbjct 787 KRHSEGTFSNDYSKYLETRRAQDF#VQWLKNSKRNGS-------LFRRHADGTYT 833 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNR 85 +||+ ||+||| | || + |++|#| || + | Sbjct 825 RRHADGTYTSDVSAYLQDQAAKEF#VSWLKTGRGRR 859 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ||||+|||++||||||++||||||#|+||||+| | + +||||||+| Sbjct 48 KRHSEGTFSNDYSKYLETRRAQDF#VEWLMNSKSNGGSA-------KRHAEGTYT 94 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWL 78 |||++||+||| | +| + ||+|#| || Sbjct 86 KRHAEGTYTSDISSFLRDQAAQNF#VAWL 113 |
| Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ||||+|||++||||| + | ||||#||||||+||+ +|||+||+| Sbjct 50 KRHSEGTFSNDYSKYQEERMAQDF#VQWLMNSKRS-------GAPSKRHADGTYT 96 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNI--AKRHDEFERHAEGTFT 104 |||+ ||+||| | || + |+||#| || + + | + ++ || +|+|| Sbjct 88 KRHADGTYTSDVSTYLQDQAAKDF#VSWLKSGRARRESAEESRNGPMSRRHVDGSFT 143 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHD 93 +|| |+|||| +| ||| |+++#+ |+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSLAAKEY#LLWVMTSKTSGKSNKRQED 177 |
| Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 || |+|||++ |||| + | |+||#+ ||||+||+ +|||+||+| Sbjct 50 KRQSEGTFSNYYSKYQEERMARDF#LHWLMNSKRS-------GAPSKRHADGTYT 96 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHD--EFERHAEGTFT 104 |||+ ||+||| | || + |+||#| || + | + + + || +|+|| Sbjct 88 KRHADGTYTSDVSTYLQDQAAKDF#VSWLKSGPARRESAEESMNGPMSRRHVDGSFT 143 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHD 93 +|| |+|||| +| ||| |+++#+ |+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSLAAKEY#LLWVMTSKTSGKSNKRQED 177 |
| Heloderma horridum | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMN 80 |||| |||||| || ++ + |#++|| | Sbjct 46 KRHSDGTFTSDLSKQMEEEAVRLF#IEWLKN 75 |
| Mus musculus | Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHA 99 |+++||| |||| +| | |||#| ||+ + +++ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDF#VNWLLAQRGKKSD--WKHNITQREA 88 |
| Protopterus dolloi | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 | |||||||| |||+| |+++ | | #|| ||+||| ++|||+| |||||||+| Sbjct 28 EAKRHSQGTFMSDYAKLLEAKHALDL#VQRLMSTKRT-GAVSKRHNEIERHAEGTYT 82 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFE-RHAEGTFT 104 || +||++||+||| | ||+ + +|#++||| + |+ +| + |||+|| | Sbjct 71 NEIERHAEGTYTSDISSYLEGQAVNEF#IRWLMKGRGRRDFSETDIEEMDRRHADGTVT 128 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 |+||+ || ||| + |+| ++|#+ ||+| | Sbjct 119 DRRHADGTVTSDINSVLESIATREF#LNWLINAK 151 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 |+ |+|||| +| ||+ |++|#+ ||++|| Sbjct 1 HADGSFTSDINKVLDTIAAKEF#LNWLISTK 30 |
| Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | Query 47 MNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ++ + ||+ | ||||||+ | |+ +#++ |+ || ++|++ +||++ || Sbjct 75 VSRNARHADGVFTSDYSRLLGQISAKKY#LESLIG-KRISSSISEDPVPIKRHSDAVFT 131 |
| Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 |||||+|||||||+|| ++|+||#| ||+|||| + + +||++|+|| Sbjct 43 RHSQGSFTSDYSKHLDVKQAKDF#VTWLLNTKRRGVDAQAGANLEKRHSDGSFT 95 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKR 83 | +|||| |+||+| + || |++|#++|| | Sbjct 84 NLEKRHSDGSFTNDMNVMLDRMSAKNF#LEWLKQQGR 119 |
| Ambloplites rupestris | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 |||||||+||+ ||+ |+||||#++|||| | |+ |++ |||+|||| Sbjct 1 HSQGTFTNDYTNYLEDRQAQDF#IRWLMNNK--RSGAAEK-----RHADGTFT 45 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDE--FERHAEGTFT 104 +|||+ |||| | | + +||#| | + + | | | || ||+|| Sbjct 36 EKRHADGTFTDDASSDFYDQAIKDF#VAKLKSGQDRREPETDRRREAFSRRHVEGSFT 92 |
| Arvicanthis ansorgei | Query 47 MNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ++ + ||+ | ||||||+ | |+ +#++ |+ || ++| + +||++ || Sbjct 58 VSRNARHADGVFTSDYSRLLGQISAKKY#LESLIG-KRISSSILEDPVPIKRHSDAVFT 114 |
| synthetic construct | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRN---NIAKRHDEFERHAEGTFT 104 +| +||++|||||| | ||+ + |++|#+ ||+ + |+ +| + |||+|+|+ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQAAKEF#IAWLVKGRGRRDFPEEVAIVEELGRRHADGSFS 152 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 +||+ |+|+ + + ||+ |+||#+ ||+ || Sbjct 144 RRHADGSFSDEMNTILDNLAARDF#INWLIQTK 175 |
| Phodopus sungorus | Query 56 GTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRN---NIAKRHDEFERHAEGTFT 104 |||||| | ||+ + |++|#+ ||+ + |+ + + |||+|+|+ Sbjct 1 GTFTSDVSSYLEGQAAKEF#IAWLVKGRGRRDFPEEVTIVEELGRRHADGSFS 52 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 +||+ |+|+ + + ||| +||#+ ||+ || Sbjct 44 RRHADGSFSDEMNTILDSLATRDF#INWLIQTK 75 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | Query 46 QMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 +++ ||||+|||++||||||+ |+||+|#|+|||| | |+ +|++ |||+|||| Sbjct 46 ELSNMKRHSEGTFSNDYSKYLEDRKAQEF#VRWLMNNK--RSGVAEK-----RHADGTFT 97 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWL 78 +|||+ |||||| | || + +||#| | Sbjct 88 EKRHADGTFTSDVSSYLKDQAIKDF#VDRL 116 |
| Sus scrofa | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ||| ||||||||||||||||||||||||||#||||||||||+||||||||||||||||||| Sbjct 45 DQMTEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRN---NIAKRHDEFERHAEGTFT 104 +| +||++|||||| | ||+ + |++|#+ ||+ + |+ + + |||+|+|+ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQAAKEF#IAWLVKGRGRRDFPEEVTIVEELRRRHADGSFS 152 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 +||+ |+|+ + + ||+ +||#+ ||++|| Sbjct 144 RRHADGSFSDEMNTVLDNLATRDF#INWLLHTK 175 |
| Neoceratodus forsteri | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 | |||||||| |||+| |++| | ||#|| ||||||| ++||| | |||||||+| Sbjct 20 EAKRHSQGTFMSDYAKLLEARHALDF#VQRLMNTKRN-GGVSKRHSEIERHAEGTYT 74 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEF-ERHAEGTFT 104 +| +||++||+||| | ||+ + | +|#|+||+ + |+ +| |||+|| | Sbjct 63 SEIERHAEGTYTSDISSYLEGQAANEF#VRWLLKGRGRRDFSDTDAEEMGRRHADGTIT 120 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 +||+ || |+| + |+| ++|#+ ||+|+| Sbjct 112 RRHADGTITNDMNNVLESIATREF#LNWLINSK 143 |
| Canis lupus familiaris | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 ||||||||||||||||||||||||||||||#||||||||||+||||||||||||||||||| Sbjct 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRN---NIAKRHDEFERHAEGTFT 104 +| +||++|||||| | ||+ + |++|#+ ||+ + |+ +| + |||+|+|+ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQAAKEF#IAWLVKGRGRRDFPEEVAIVEEFRRRHADGSFS 152 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 +||+ |+|+ + + ||+ +||#+ ||+ || Sbjct 144 RRHADGSFSDEMNTVLDTLATRDF#INWLLQTK 175 |
| Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 |||||||+|||||||+||||||#|||||+|||+ | ||+||+| Sbjct 1 HSQGTFTNDYSKYLDTRRAQDF#VQWLMSTKRS-GGITXXXXXXXXHADGTYT 51 Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNR 85 |+ ||+||| | || + |+ |#| || + | Sbjct 45 HADGTYTSDVSSYLQDQAAKKF#VTWLKQGQDRR 77 |
| Hydrolagus colliei | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 |||+ | |+||||||||+|| +||#||||++|||| | |+ +|||+| +| Sbjct 30 KRHTDGIFSSDYSKYLDNRRTKDF#VQWLLSTKRNGANT----DKTKRHADGIYT 79 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNN--IAKRHDEFERHAE 100 ++ |||+ | +||| + | +++ |#|+ | | + +|+ ++||| | | | Sbjct 68 DKTKRHADGIYTSDVASLTDYLKSKRF#VESLSNYNKRQNDRRMSKRHVEGARRTE 122 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFE 96 | | ++ + +|+| ||+++ |+||#+ ||+ + | + |+ | | Sbjct 116 EGARRTEEDYPNDFSSYLEAKAAKDF#IDWLIK-GQGRRDFAEESREIE 162 |
| Heloderma suspectum, venom, Peptide, 39 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMN 80 | +|||||| || ++ + |#++|| | Sbjct 1 HGEGTFTSDLSKQMEEEAVRLF#IEWLKN 28 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | Query 56 GTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRN 86 ||+||| | || + |++|#| || + |+ Sbjct 3 GTYTSDVSSYLQDQAAKEF#VSWLKTGRGRRD 33 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNR 85 |+ || ||| | +|+ + ++|#+ ||++ + | Sbjct 1 HADGTLTSDISSFLEKQATKEF#IAWLVSGRGRR 33 |
| Agkistrodon piscivorus | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRH---DEFERHAEGTFT 104 | +||+ ||+||| | ||+ + |++|#+ ||+| + |+ | | |||+|||| Sbjct 32 EYERHADGTYTSDISSYLEGQAAKEF#IAWLVNGRGRRDFSEGAHTGEDLGRRHADGTFT 90 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNI 88 +||+ |||||||+| || |+|#++||+| | + ++ Sbjct 82 RRHADGTFTSDYNKLLDDIATQEF#LKWLINQKVTQRDL 119 Query 66 LDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 |+ | ++|#+ | + | +|+ | |+||||+||+| Sbjct 9 LEDREKENF#LDQLAS-----NGLARPHAEYERHADGTYT 42 |
[Site 2] SDYSKYLDSR69-RAQDFVQWLM
Arg69  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser60 | Asp61 | Tyr62 | Ser63 | Lys64 | Tyr65 | Leu66 | Asp67 | Ser68 | Arg69 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg70 | Ala71 | Gln72 | Asp73 | Phe74 | Val75 | Gln76 | Trp77 | Leu78 | Met79 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PLSDPDQMNEDKRHSQGTFTSDYSKYLDSRRAQDFVQWLMNTKRNRNNIAKRHDEFERHA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 130.00 | 10 | glucagon, isoform CRA_b |
| 2 | Pan troglodytes | 102.00 | 2 | PREDICTED: fibroblast activation protein, alpha su |
| 3 | N/A | 79.70 | 44 | 0 |
| 4 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | 64.70 | 1 | AAB31092.1 |
| 5 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | 61.60 | 1 | glucagon=proglucagon-derived 3.5 kda peptide |
| 6 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | 60.80 | 1 | glucagon II |
| 7 | Contains: Glucagon; Glucagon-like peptide | 60.10 | 4 | GLUC_AMICA Glucagon precursor |
| 8 | Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | 60.10 | 1 | glucagon |
| 9 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | 58.50 | 1 | glucagon G-29 |
| 10 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | 57.80 | 1 | GLUC1_ORENI Glucagon-1 (Glucagon I) gb |
| 11 | Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | 56.20 | 1 | GLUC_LAMFL Glucagon gb |
| 12 | deshis1, Desphe6, Glu9 | 53.50 | 1 | A Chain A, Nmr Solution Structure Of The Glucagon |
| 13 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | 53.50 | 1 | glucagon |
| 14 | Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | 47.00 | 1 | GLUC1_PETMA Glucagon-1 precursor |
| 15 | Agkistrodon piscivorus | 45.10 | 1 | proglucagon |
| 16 | Gallus gallus | 44.30 | 4 | glucagon isoform 1 |
| 17 | Meleagris gallopavo | 44.30 | 2 | preproglucagon B |
| 18 | Monodelphis domestica | 43.90 | 3 | PREDICTED: similar to Gastric inhibitory polypepti |
| 19 | Oncorhynchus tshawytscha | 43.50 | 1 | S78474_1 proglucagon |
| 20 | Rattus norvegicus | 42.70 | 5 | Glucose-dependent insulinotropic peptide |
| 21 | Canis familiaris | 42.70 | 2 | PREDICTED: similar to Gastric inhibitory polypepti |
| 22 | Xenopus laevis | 42.40 | 5 | proglucagon I |
| 23 | Xenopus tropicalis | 42.40 | 2 | glucagon |
| 24 | Danio rerio | 42.00 | 5 | hypothetical protein LOC494052 |
| 25 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | 42.00 | 1 | GLUC_HELSU Glucagon precursor |
| 26 | Heloderma suspectum | 41.60 | 2 | proglucagon |
| 27 | Hoplobatrachus rugulosus | 41.60 | 1 | AF324209_1 proglucagon |
| 28 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | 41.20 | 1 | GLUC2_LOPAM Glucagon-2 precursor (Glucagon II) |
| 29 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | 41.20 | 1 | glucagon-like peptide, GLP |
| 30 | Macaca mulatta | 40.80 | 3 | PREDICTED: gastric inhibitory polypeptide |
| 31 | Bos taurus | 40.40 | 5 | PREDICTED: similar to Gastric inhibitory polypepti |
| 32 | Ictalurus punctatus | 40.40 | 3 | proglucagon I |
| 33 | Protopterus dolloi | 40.40 | 1 | proglucagon |
| 34 | Sebastes caurinus | 40.00 | 4 | proglucagon II |
| 35 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | 40.00 | 2 | GLUC_CARAU Glucagon precursor |
| 36 | Tetraodon nigroviridis | 40.00 | 2 | unnamed protein product |
| 37 | Heloderma horridum | 40.00 | 1 | EXE3_HELHO Exendin-3 precursor emb |
| 38 | Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC1_ONCMY Glucagon-1 precursor (Glucagon I) |
| 39 | Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC2_ONCMY Glucagon-2 precursor (Glucagon II) |
| 40 | Mus musculus | 39.70 | 6 | gastric inhibitory polypeptide |
| 41 | Neoceratodus forsteri | 39.70 | 1 | proglucagon |
| 42 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | 38.90 | 1 | glucagon-like peptide-2, GLP-2=proglucagon-derived |
| 43 | Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | 38.10 | 1 | GLUC2_PETMA Glucagon-2 precursor |
| 44 | synthetic construct | 37.00 | 1 | Homo sapiens glucagon |
| 45 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | 36.60 | 1 | GLUC1_LOPAM Glucagon-1 precursor (Glucagon I) |
| 46 | Phodopus sungorus | 36.60 | 1 | glucagon precursor |
| 47 | Sus scrofa | 36.20 | 1 | glucagon |
| 48 | Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | 35.80 | 1 | GLUC_LEPSP Glucagon precursor |
| 49 | Canis lupus familiaris | 35.80 | 1 | glucagon |
| 50 | Squalus acanthias | 35.40 | 1 | proglucagon |
| 51 | Hydrolagus colliei | 35.40 | 1 | proglucagon |
| 52 | Heloderma suspectum, venom, Peptide, 39 aa | 35.40 | 1 | A Chain A, Solution Structure Of Exendin-4 In 30-V |
| 53 | Bufo marinus | 35.40 | 1 | proglucagon |
| 54 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | 34.70 | 1 | glucagon-like peptide-1, GLP-1=proglucagon-derived |
| 55 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | 34.70 | 1 | GLUC2_ORENI Glucagon-2 (Glucagon II) gb |
| 56 | Capra hircus | 34.30 | 1 | glucagon |
| 57 | Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | 33.50 | 1 | VIP_MOUSE VIP peptides precursor |
| 58 | Arvicanthis ansorgei | 32.70 | 1 | vasoactive intestinal polypeptide |
| 59 | Candida glabrata | 32.70 | 1 | unnamed protein product |
| 60 | Ambloplites rupestris | 32.30 | 1 | proglucagon |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 |
| Pan troglodytes | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDE 94 ||||||||||||||||||||||||||||||#||||||||||||||| +++| || | Sbjct 962 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN-SSVAYRHAE 1015 |
| N/A | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIA 89 |||||||||||||||||#|||||||||||||||+|||| Sbjct 1 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIA 37 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN 84 ||+||||||||||+|+|#||+|||||||+|||| Sbjct 1 HSEGTFTSDYSKYMDNR#RAKDFVQWLMSTKRN 32 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 |||||||||||||||+|#|||||+||||+| Sbjct 1 HSQGTFTSDYSKYLDNR#RAQDFIQWLMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN 84 ||+|||++||||||++|#||||||||| |+||| Sbjct 1 HSEGTFSNDYSKYLETR#RAQDFVQWLKNSKRN 32 |
| Contains: Glucagon; Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 |||||||+|||||+|+|#||||||||||+| Sbjct 1 HSQGTFTNDYSKYMDTR#RAQDFVQWLMST 29 |
| Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 |||||||+|||||+|+|#||||||||||+| Sbjct 1 HSQGTFTNDYSKYMDTR#RAQDFVQWLMST 29 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 ||+||||||||||+|+|#||+|||||||+| Sbjct 1 HSEGTFTSDYSKYMDNR#RAKDFVQWLMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN 84 ||+|||++||||||+ |#+|||||+|||| ||+ Sbjct 1 HSEGTFSNDYSKYLEDR#KAQDFVRWLMNNKRS 32 |
| Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 ||||+|||||||||||+#+|+||| ||||| Sbjct 1 HSQGSFTSDYSKYLDSK#QAKDFVIWLMNT 29 |
| deshis1, Desphe6, Glu9 | Query 54 SQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 |||| ||+||||||||#|||||||||||| Sbjct 1 SQGT-TSEYSKYLDSR#RAQDFVQWLMNT 27 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMN 80 ||+|||||||||||+++#+|+|||+|||| Sbjct 1 HSEGTFTSDYSKYLENK#QAKDFVRWLMN 28 |
| Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRH 92 ||||+|||||||||||+++#+|+|||+|||| || + + +|| Sbjct 41 KRHSEGTFTSDYSKYLENK#QAKDFVRWLMNAKRGGSELQRRH 82 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNR---------NNIAKRHDE 94 +||+ ||||+| + |||++# |+||| || + ++| |+ ||| | Sbjct 80 RRHADGTFTNDMTSYLDAK#AARDFVSWLARSDKSRRDGGDHLAENSEDKRHAE 132 |
| Agkistrodon piscivorus | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNI 88 +||+ |||||||+| || # |+|++||+| | + ++ Sbjct 82 RRHADGTFTSDYNKLLDDI#ATQEFLKWLINQKVTQRDL 119 Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRH---DEFERHA 99 | +||+ ||+||| | ||+ +# |++|+ ||+| + |+ | | ||| Sbjct 32 EYERHADGTYTSDISSYLEGQ#AAKEFIAWLVNGRGRRDFSEGAHTGEDLGRRHA 85 |
| Gallus gallus | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN--------------- 84 || + |+|| |||||||||||||||||||#||||||||||+|||| Sbjct 42 PLDESRQLNEVKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMSTKRNGQQGQEDKENDKFPD 101 Query 85 ---RNNIAKRHDEFERHA 99 | |+||| |||||| Sbjct 102 QLSSNAISKRHSEFERHA 119 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNI 88 +||+ |||||| +| || # |++|++||+||| + ++ Sbjct 164 RRHADGTFTSDINKILDDM#AAKEFLKWLINTKVTQRDL 201 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRN 86 +| +||++||+||| + ||+ +# |++|+ ||+| + |+ Sbjct 113 SEFERHAEGTYTSDITSYLEGQ#AAKEFIAWLVNGRGRRD 151 |
| Meleagris gallopavo | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN--------------- 84 || + |+|| |||||||||||||||||||#||||||||||+|||| Sbjct 42 PLDESRQLNEVKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMSTKRNGQQGQEDKENDEFPD 101 Query 85 ---RNNIAKRHDEFERHA 99 | |+||| |||||| Sbjct 102 QLSSNAISKRHSEFERHA 119 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNI 88 +||+ |||||| +| || # |++|++||+||| + ++ Sbjct 164 RRHADGTFTSDINKILDDM#AAKEFLKWLINTKVTQRDL 201 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRN 86 +| +||++||+||| + ||+ +# |++|+ ||+| + |+ Sbjct 113 SEFERHAEGTYTSDITSYLEGQ#AAKEFIAWLVNGRGRRD 151 |
| Monodelphis domestica | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 +|+++||| |||| +| # |||| ||++ | +| + ||+ || | Sbjct 51 QRYAEGTFISDYSITMDKI#MQQDFVNWLLSQKGKKN--SWRHNITERKA 97 |
| Oncorhynchus tshawytscha | Query 54 SQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRH 92 | |||++||||| + |# ||||||||||+||+ +||| Sbjct 2 SXGTFSNDYSKYQEER#MAQDFVQWLMNSKRS-GAPSKRH 39 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNR 85 |||+ ||+||| | || +# |+||| || + + | Sbjct 37 KRHADGTYTSDVSTYLQDQ#AAKDFVSWLKSGRARR 71 |
| Rattus norvegicus | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 |+++||| |||| +| #| |||| ||+ | +|+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKI#RQQDFVNWLLAQKGKKND--WKHNLTQREA 88 |
| Canis familiaris | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 |+++||| |||| +| #| |||| ||+ | +|+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKI#RQQDFVNWLLAQKGKKND--WKHNITQREA 88 |
| Xenopus laevis | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 + | +|+ | |||||||||||||||||||#||||||||||||||+ +++|+ ++|||| Sbjct 41 VDDSEQLKEVKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRS-GELSRRNADYERHA 98 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 +||++|||||| ++ || +# |++|+ ||+| ++ |++|+ |||||| Sbjct 95 ERHAEGTFTSDVTQQLDEK#AAKEFIDWLINGGPSKEIISRRNAEFERHA 143 Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLM--NTKRNRNNIAKRHDEFERHA 99 ||++||||+| + ||+ +# |++|| ||+ ||| + + + ||| Sbjct 179 RHAEGTFTNDMTNYLEEK#AAKEFVGWLIKGRPKRNFSEVHSVEEMDRRHA 228 Query 50 DKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNI 88 |+||+ |+||+| +| || # ||+|+ |++||+ ++ Sbjct 224 DRRHADGSFTNDINKVLDII#AAQEFLDWVINTQETERDL 262 Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDE 94 | +||++||+|+| ++||+ +# |++|++||+ | + + || | Sbjct 138 EFERHAEGTYTNDVTEYLEEK#AAKEFIEWLIKGKPKKIRYS-RHAE 182 |
| Xenopus tropicalis | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 + | |+ | |||||||||||||||||||#|||||+||||||||+ +++|+ ++|||| Sbjct 41 VDDSQQLKEVKRHSQGTFTSDYSKYLDSR#RAQDFIQWLMNTKRS-GGLSRRNADYERHA 98 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 +||++|||||| +++|| +# |++|+ ||+| ++ |++|+ + |||| Sbjct 95 ERHAEGTFTSDVTQHLDEK#AAKEFIDWLINGGPSKEIISRRNADVERHA 143 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDE 94 +||++||+|+| ++||+ +# |++|++||+| | + + || | Sbjct 140 ERHAEGTYTNDVTEYLEEK#AAKEFIEWLINGKPKKIRYS-RHAE 182 Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRN 86 ||++||||+| + ||+ +# |++|| ||+ + || Sbjct 179 RHAEGTFTNDMTNYLEEK#AAKEFVGWLIKGRPKRN 213 Query 50 DKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 |+||+ |+||+| +| || # ||+|+ |++||+ Sbjct 224 DRRHADGSFTNDINKVLDII#AAQEFLDWVINTQ 256 |
| Danio rerio | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDE 94 ||||+|||++||||||++|#||||||||||| ||| ++ ||| | Sbjct 48 KRHSEGTFSNDYSKYLETR#RAQDFVQWLMNAKRNGGSV-KRHAE 90 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWL 78 |||++||+||| | || +# || || || Sbjct 86 KRHAEGTYTSDVSSYLQDQ#AAQSFVAWL 113 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKR---------------- 83 || | |+|| |||||||||||||||||+|#|||||||||||||| Sbjct 40 PLDDSRQLNEVKRHSQGTFTSDYSKYLDTR#RAQDFVQWLMNTKRSGQQGVEEREKENLLD 99 Query 84 --NRNNIAKRHDEFERHA 99 + | +|+ | |+|||| Sbjct 100 QLSSNGLARHHAEYERHA 117 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNI 88 +||+ |||||||++ || # |+|++||+| | + ++ Sbjct 162 RRHADGTFTSDYNQLLDDI#ATQEFLKWLINQKVTQRDL 199 Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRN 86 | +||+ | +||| | ||+ +# |++|+ ||+| + |+ Sbjct 112 EYERHADGRYTSDISSYLEGQ#AAKEFIAWLVNGRGRRD 149 |
| Heloderma suspectum | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKR---------------- 83 || | |+|| |||||||||||||||||+|#|||||||||||||| Sbjct 40 PLDDSRQLNEVKRHSQGTFTSDYSKYLDTR#RAQDFVQWLMNTKRSGQQGVEEREKENLLD 99 Query 84 --NRNNIAKRHDEFERHA 99 + | +|+ | |+|||| Sbjct 100 QLSSNGLARHHAEYERHA 117 Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNR 85 | +||+ | +||| | ||+ +# |++|+ ||+| + | Sbjct 112 EYERHADGRYTSDISSYLEGQ#AAKEFIAWLVNGRGRR 148 |
| Hoplobatrachus rugulosus | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 | | | || |||||||||||||||||||#|||||||||||+||+ |+||+ +||||| Sbjct 41 LDDSDHHNEVKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNSKRS-GGISKRNVQFERHA 98 Query 50 DKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 |+||+ |+||||++| || +# ||+|+ |++|| Sbjct 178 DRRHADGSFTSDFNKALDIK#AAQEFLDWIINT 209 Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRN 86 ||++|||||| + ||+ +# |++|| ||+ + || Sbjct 134 RHAEGTFTSDMTSYLEEK#AAKEFVDWLIKGRPKRN 168 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDE 94 +||++||+|+| +++|+ +# |++|+ ||+ | + ++ || | Sbjct 95 ERHAEGTYTNDVTQFLEEK#AAKEFIDWLIKGKPKKQRLS-RHAE 137 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRH 92 |++| + ||||+|||++||||||++|#||||||||| |+| || + +|| Sbjct 40 LTEPIEPLNMKRHSEGTFSNDYSKYLETR#RAQDFVQWLKNSK--RNGLFRRH 89 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWL 78 +||+ ||+||| | || +# |+||| || Sbjct 87 RRHADGTYTSDVSSYLQDQ#AAKDFVSWL 114 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN 84 |+ ||||+| + |||++# |+||| || + ++ Sbjct 1 HADGTFTNDMTSYLDAK#AARDFVSWLARSDKS 32 |
| Macaca mulatta | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 |+++||| |||| +| # |||| ||+ | +|+ +|+ +| | Sbjct 51 RYAEGTFISDYSIAMDKI#HQQDFVNWLLAQKGKKND--WKHNITQREA 96 |
| Bos taurus | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNN 87 |+++||| |||| +| #| |||| ||+ | +++ Sbjct 81 RYAEGTFISDYSIAMDKI#RQQDFVNWLLAQKGKKSD 116 |
| Ictalurus punctatus | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRH 92 +|||+|||++||||||++|#|||||||||||+||+ ++ |+|| Sbjct 49 RRHSEGTFSNDYSKYLETR#RAQDFVQWLMNSKRS-DSPARRH 89 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRH 92 +|| |+|||| +| ||| # |++++||+||+ + + ||| Sbjct 134 RRHVDGSFTSDVNKVLDSL#AAKEYLQWVMNSPSSAS--GKRH 173 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWL 78 +||+ ||+||| | || +# |+||+ || Sbjct 87 RRHADGTYTSDVSSYLQDQ#AAKDFITWL 114 |
| Protopterus dolloi | Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 | |||||||| |||+| |+++# | | || ||+||| ++|||+| |||| Sbjct 28 EAKRHSQGTFMSDYAKLLEAK#HALDLVQRLMSTKRT-GAVSKRHNEIERHA 77 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFER 97 || +||++||+||| | ||+ +# +|++||| + |+ +| +| Sbjct 71 NEIERHAEGTYTSDISSYLEGQ#AVNEFIRWLMKGRGRRDFSETDIEEMDR 120 Query 42 SDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 |+ | |+||+ || ||| + |+| # ++|+ ||+| | Sbjct 111 SETDIEEMDRRHADGTVTSDINSVLESI#ATREFLNWLINAK 151 |
| Sebastes caurinus | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRH 92 |++| ++ ||||+|||++||||||++|#||||||||| |+||| ++ +|| Sbjct 40 LAEPIELPNMKRHSEGTFSNDYSKYLETR#RAQDFVQWLKNSKRN-GSLFRRH 90 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNR 85 +||+ ||+||| | || +# |++|| || + | Sbjct 88 RRHADGTYTSDVSSYLQDQ#AAKEFVYWLKTGRGRR 122 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDE 94 ||||+|||++||||||++|#||||||+||||+| | + |||| | Sbjct 48 KRHSEGTFSNDYSKYLETR#RAQDFVEWLMNSKSNGGS-AKRHAE 90 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWL 78 |||++||+||| | +| +# ||+|| || Sbjct 86 KRHAEGTYTSDISSFLRDQ#AAQNFVAWL 113 |
| Tetraodon nigroviridis | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRH 92 |++| ++ ||||+|||++||||||++|#||||||||| |+||| ++ +|| Sbjct 777 LTEPSDLSNMKRHSEGTFSNDYSKYLETR#RAQDFVQWLKNSKRN-GSLFRRH 827 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNR 85 +||+ ||+||| | || +# |++|| || + | Sbjct 825 RRHADGTYTSDVSAYLQDQ#AAKEFVSWLKTGRGRR 859 |
| Heloderma horridum | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMN 80 |||| |||||| || ++ # + |++|| | Sbjct 46 KRHSDGTFTSDLSKQMEEE#AVRLFIEWLKN 75 |
| Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRH 92 | | ++ ||||+|||++||||| + |# ||||||||||+||+ +||| Sbjct 40 LEDLMGVSNVKRHSEGTFSNDYSKYQEER#MAQDFVQWLMNSKRS-GAPSKRH 90 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNN 87 |||+ ||+||| | || +# |+||| || + + | + Sbjct 88 KRHADGTYTSDVSTYLQDQ#AAKDFVSWLKSGRARRES 124 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHD 93 +|| |+|||| +| ||| # |++++ |+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSL#AAKEYLLWVMTSKTSGKSNKRQED 177 |
| Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRH 92 || |+|||++ |||| + |# |+||+ ||||+||+ +||| Sbjct 50 KRQSEGTFSNYYSKYQEER#MARDFLHWLMNSKRS-GAPSKRH 90 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNN 87 |||+ ||+||| | || +# |+||| || + | + Sbjct 88 KRHADGTYTSDVSTYLQDQ#AAKDFVSWLKSGPARRES 124 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHD 93 +|| |+|||| +| ||| # |++++ |+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSL#AAKEYLLWVMTSKTSGKSNKRQED 177 |
| Mus musculus | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 |+++||| |||| +| #| |||| ||+ + +++ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKI#RQQDFVNWLLAQRGKKSD--WKHNITQREA 88 |
| Neoceratodus forsteri | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 |+ + | |||||||| |||+| |++|# | |||| ||||||| ++||| | |||| Sbjct 11 PVDASREHTEAKRHSQGTFMSDYAKLLEAR#HALDFVQRLMNTKRN-GGVSKRHSEIERHA 69 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRN 86 +| +||++||+||| | ||+ +# | +||+||+ + |+ Sbjct 63 SEIERHAEGTYTSDISSYLEGQ#AANEFVRWLLKGRGRRD 101 Query 42 SDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 || | +||+ || |+| + |+| # ++|+ ||+|+| Sbjct 103 SDTDAEEMGRRHADGTITNDMNNVLESI#ATREFLNWLINSK 143 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 |+ |+|||| +| ||+ # |++|+ ||++|| Sbjct 1 HADGSFTSDINKVLDTI#AAKEFLNWLISTK 30 |
| Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN------RNNIAKRHDE 94 |||||+|||||||+|| +#+|+||| ||+|||| |+ ||| + Sbjct 43 RHSQGSFTSDYSKHLDVK#QAKDFVTWLLNTKRRGVDAQAGANLEKRHSD 91 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKR 83 | +|||| |+||+| + || # |++|++|| | Sbjct 84 NLEKRHSDGSFTNDMNVMLDRM#SAKNFLEWLKQQGR 119 |
| synthetic construct | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 +||+ |+|+ + + ||+ # |+||+ ||+ || Sbjct 144 RRHADGSFSDEMNTILDNL#AARDFINWLIQTK 175 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | Query 43 DPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRH 92 +| +++ ||||+|||++||||||+ |#+||+||+|||| ||+ ||| Sbjct 43 EPRELSNMKRHSEGTFSNDYSKYLEDR#KAQEFVRWLMNNKRS-GVAEKRH 91 Query 50 DKRHSQGTFTSDYSKYLDSR#RAQDFVQWL 78 +|||+ |||||| | || +# +||| | Sbjct 88 EKRHADGTFTSDVSSYLKDQ#AIKDFVDRL 116 |
| Phodopus sungorus | Query 56 GTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRN 86 |||||| | ||+ +# |++|+ ||+ + |+ Sbjct 1 GTFTSDVSSYLEGQ#AAKEFIAWLVKGRGRRD 31 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 +||+ |+|+ + + ||| # +||+ ||+ || Sbjct 44 RRHADGSFSDEMNTILDSL#ATRDFINWLIQTK 75 |
| Sus scrofa | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 || ||||| |||||||||||||||||||||#|||||||||||||||+|||||||||||||| Sbjct 40 PLDDPDQMTEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 +||+ |+|+ + + ||+ # +||+ ||++|| Sbjct 144 RRHADGSFSDEMNTVLDNL#ATRDFINWLLHTK 175 |
| Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN 84 |||||||+|||||||+|#||||||||||+|||+ Sbjct 1 HSQGTFTNDYSKYLDTR#RAQDFVQWLMSTKRS 32 Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNR 85 |+ ||+||| | || +# |+ || || + | Sbjct 45 HADGTYTSDVSSYLQDQ#AAKKFVTWLKQGQDRR 77 |
| Canis lupus familiaris | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 ||+| |||||||||||||||||||||||||#|||||||||||||||+|||||||||||||| Sbjct 40 PLNDLDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRN 86 +| +||++|||||| | ||+ +# |++|+ ||+ + |+ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQ#AAKEFIAWLVKGRGRRD 131 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 +||+ |+|+ + + ||+ # +||+ ||+ || Sbjct 144 RRHADGSFSDEMNTVLDTL#ATRDFINWLLQTK 175 |
| Squalus acanthias | Query 44 PDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFE 96 |++| |+||+ |||||+ + ||+ # |++|+ |++|+| | | | | Sbjct 118 PEEM--DRRHADGTFTSEINIVLDTI#AAKEFLNWILNSK----TIQSRDSEIE 164 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWL--MNTKRNRNNIAKRHDEFERH 98 ++ |||++||+||| | #+|+ || | | ++| +|+||| | || Sbjct 20 DKTKRHAEGTYTSDVDSLSDYF#KAKRFVDSLTSYNKRQNGRSISKRHVESTRH 72 Query 67 DSR#RAQDFVQWLMNTKRNRNNIAKRHDE 94 |+|#||+|||||||+|||| + ||| | Sbjct 1 DNR#RAKDFVQWLMSTKRNGDK-TKRHAE 27 Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNR 85 | ||++|++ | | ||+++# |+||+ ||+ + | Sbjct 68 ESTRHTEGSYR-DISSYLEAK#AAKDFINWLIKGRGRR 103 |
| Hydrolagus colliei | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 +|+| + + |||+ | |+||||||||+|#| +||||||++|||| | |+ +||| Sbjct 20 VSNPSTVTDVKRHTDGIFSSDYSKYLDNR#RTKDFVQWLLSTKRNGANT----DKTKRHA 74 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNN--IAKRHDEFER 97 ++ |||+ | +||| + | #+++ ||+ | | + +|+ ++||| | | Sbjct 68 DKTKRHADGIYTSDVASLTDYL#KSKRFVESLSNYNKRQNDRRMSKRHVEGAR 119 Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFE 96 | | ++ + +|+| ||+++# |+||+ ||+ + | + |+ | | Sbjct 116 EGARRTEEDYPNDFSSYLEAK#AAKDFIDWLIK-GQGRRDFAEESREIE 162 |
| Heloderma suspectum, venom, Peptide, 39 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMN 80 | +|||||| || ++ # + |++|| | Sbjct 1 HGEGTFTSDLSKQMEEE#AVRLFIEWLKN 28 |
| Bufo marinus | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLM--NTKRNRNNIAKRHDEFERHA 99 ||++|||||| + +|+ +# |++|| ||+ ||| ++++ | ||| Sbjct 62 RHAEGTFTSDMTSFLEEK#AAKEFVDWLIKGRPKRNFSDVSAADDMDRRHA 111 Query 50 DKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 |+||+ |+||||++| || +# ||+|+ |++|| Sbjct 107 DRRHADGSFTSDFNKALDIK#AAQEFLDWIINT 138 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDE 94 +||++||+|+| +++|+ +# |++|+ ||+ + ++ || | Sbjct 23 ERHAEGTYTNDVTQFLEEK#AAKEFIDWLLKGIPKKQRLS-RHAE 65 Query 73 DFVQWLMNTKRNRNNIAKRHDEFERHA 99 || |||||+||+ +++|+ +||||| Sbjct 1 DFAQWLMNSKRS-GGMSRRNVQFERHA 26 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNR 85 |+ || ||| | +|+ +# ++|+ ||++ + | Sbjct 1 HADGTLTSDISSFLEKQ#ATKEFIAWLVSGRGRR 33 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | Query 56 GTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRN 86 ||+||| | || +# |++|| || + |+ Sbjct 3 GTYTSDVSSYLQDQ#AAKEFVSWLKTGRGRRD 33 |
| Capra hircus | Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNR 85 +| +||++|||||| | ||+ +# |++|+ ||+ + | Sbjct 8 DEFERHAEGTFTSDVSSYLEGQ#AAKEFIAWLVKGRGRR 45 Query 86 NNIAKRHDEFERHA 99 |||||||||||||| Query 86 NNIAKRHDEFERHA 99 |
| Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | Query 47 MNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 ++ + ||+ | ||||||+ | # |+ +++ |+ || ++|++ +||+ Sbjct 75 VSRNARHADGVFTSDYSRLLGQI#SAKKYLESLIG-KRISSSISEDPVPIKRHS 126 |
| Arvicanthis ansorgei | Query 47 MNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 ++ + ||+ | ||||||+ | # |+ +++ |+ || ++| + +||+ Sbjct 58 VSRNARHADGVFTSDYSRLLGQI#SAKKYLESLIG-KRISSSILEDPVPIKRHS 109 |
| Candida glabrata | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAK 90 |+| | ||+ | | | | + +|+ # || |+ + | +| +| Sbjct 351 LADGDDDEEDEEDSSSTSESSYDEEIDNE#EKQDIVEEEEDADENDDNFSK 400 |
| Ambloplites rupestris | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRH 92 |||||||+||+ ||+ |#+||||++|||| ||+ ||| Sbjct 1 HSQGTFTNDYTNYLEDR#QAQDFIRWLMNNKRS-GAAEKRH 39 Query 50 DKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMN--------TKRNRNNIAKRHDE 94 +|||+ |||| | | +# +||| | + | | | ++|| | Sbjct 36 EKRHADGTFTDDASSDFYDQ#AIKDFVAKLKSGQDRREPETDRRREAFSRRHVE 88 |
[Site 3] SRRAQDFVQW77-LMNTKRNRNN
Trp77  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser68 | Arg69 | Arg70 | Ala71 | Gln72 | Asp73 | Phe74 | Val75 | Gln76 | Trp77 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu78 | Met79 | Asn80 | Thr81 | Lys82 | Arg83 | Asn84 | Arg85 | Asn86 | Asn87 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| NEDKRHSQGTFTSDYSKYLDSRRAQDFVQWLMNTKRNRNNIAKRHDEFERHAEGTFTSDV |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 127.00 | 10 | glucagon, isoform CRA_b |
| 2 | N/A | 79.70 | 45 | 0 |
| 3 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | 64.70 | 1 | AAB31092.1 |
| 4 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | 61.60 | 1 | glucagon=proglucagon-derived 3.5 kda peptide |
| 5 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | 60.80 | 1 | glucagon II |
| 6 | Contains: Glucagon; Glucagon-like peptide | 60.10 | 4 | GLUC_AMICA Glucagon precursor |
| 7 | Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | 60.10 | 1 | glucagon |
| 8 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | 58.50 | 1 | glucagon G-29 |
| 9 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | 57.80 | 1 | GLUC1_ORENI Glucagon-1 (Glucagon I) gb |
| 10 | Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | 56.20 | 1 | GLUC_LAMFL Glucagon gb |
| 11 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | 53.50 | 1 | glucagon |
| 12 | deshis1, Desphe6, Glu9 | 53.50 | 1 | A Chain A, Nmr Solution Structure Of The Glucagon |
| 13 | Monodelphis domestica | 48.90 | 3 | PREDICTED: similar to Gastric inhibitory polypepti |
| 14 | Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | 48.90 | 1 | GLUC1_PETMA Glucagon-1 precursor |
| 15 | Xenopus laevis | 47.00 | 5 | proglucagon II |
| 16 | Hoplobatrachus rugulosus | 46.60 | 1 | AF324209_1 proglucagon |
| 17 | Bufo marinus | 46.60 | 1 | proglucagon |
| 18 | Gallus gallus | 46.20 | 4 | glucagon isoform 1 |
| 19 | Meleagris gallopavo | 46.20 | 2 | preproglucagon B |
| 20 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | 44.70 | 1 | GLUC_HELSU Glucagon precursor |
| 21 | Capra hircus | 43.90 | 1 | glucagon |
| 22 | Oncorhynchus tshawytscha | 43.50 | 1 | S78474_1 proglucagon |
| 23 | Ictalurus punctatus | 43.10 | 3 | proglucagon I |
| 24 | Rattus norvegicus | 42.70 | 6 | Glucose-dependent insulinotropic peptide |
| 25 | Canis familiaris | 42.70 | 2 | PREDICTED: similar to Gastric inhibitory polypepti |
| 26 | Ambloplites rupestris | 42.70 | 1 | proglucagon |
| 27 | Pan troglodytes | 42.40 | 2 | PREDICTED: fibroblast activation protein, alpha su |
| 28 | Xenopus tropicalis | 42.40 | 2 | glucagon |
| 29 | Danio rerio | 42.00 | 6 | hypothetical protein LOC494052 |
| 30 | Bos taurus | 42.00 | 5 | vasoactive intestinal peptide |
| 31 | Macaca mulatta | 41.60 | 3 | PREDICTED: vasoactive intestinal peptide |
| 32 | Heloderma suspectum | 41.60 | 2 | proglucagon |
| 33 | Squalus acanthias | 41.60 | 1 | proglucagon |
| 34 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | 41.20 | 1 | glucagon-like peptide, GLP |
| 35 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | 41.20 | 1 | GLUC2_LOPAM Glucagon-2 precursor (Glucagon II) |
| 36 | Sebastes caurinus | 40.80 | 4 | proglucagon I |
| 37 | Tetraodon nigroviridis | 40.00 | 3 | unnamed protein product |
| 38 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | 40.00 | 2 | GLUC_CARAU Glucagon precursor |
| 39 | Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC2_ONCMY Glucagon-2 precursor (Glucagon II) |
| 40 | Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC1_ONCMY Glucagon-1 precursor (Glucagon I) |
| 41 | Heloderma horridum | 40.00 | 1 | EXE3_HELHO Exendin-3 precursor emb |
| 42 | Mus musculus | 39.70 | 6 | Gastric inhibitory polypeptide |
| 43 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | 38.90 | 1 | glucagon-like peptide-2, GLP-2=proglucagon-derived |
| 44 | Protopterus dolloi | 38.90 | 1 | proglucagon |
| 45 | Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | 38.50 | 1 | VIP_MOUSE VIP peptides precursor |
| 46 | Agkistrodon piscivorus | 38.50 | 1 | proglucagon |
| 47 | Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | 38.10 | 1 | GLUC2_PETMA Glucagon-2 precursor |
| 48 | Arvicanthis ansorgei | 37.70 | 1 | vasoactive intestinal polypeptide |
| 49 | synthetic construct | 37.00 | 1 | Homo sapiens glucagon |
| 50 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | 36.60 | 1 | GLUC1_LOPAM Glucagon-1 precursor (Glucagon I) |
| 51 | Phodopus sungorus | 36.60 | 1 | glucagon precursor |
| 52 | Neoceratodus forsteri | 36.20 | 1 | proglucagon |
| 53 | Sus scrofa | 36.20 | 1 | glucagon |
| 54 | Canis lupus familiaris | 35.80 | 1 | glucagon |
| 55 | Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | 35.80 | 1 | GLUC_LEPSP Glucagon precursor |
| 56 | Hydrolagus colliei | 35.40 | 1 | proglucagon |
| 57 | Heloderma suspectum, venom, Peptide, 39 aa | 35.40 | 1 | A Chain A, Solution Structure Of Exendin-4 In 30-V |
| 58 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | 34.70 | 1 | GLUC2_ORENI Glucagon-2 (Glucagon II) gb |
| 59 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | 34.70 | 1 | glucagon-like peptide-1, GLP-1=proglucagon-derived |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSD 106 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||| Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSD 106 |
| N/A | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIA 89 |||||||||||||||||||||||||#|||||||+|||| Sbjct 1 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIA 37 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRN 84 ||+||||||||||+|+|||+|||||#||+|||| Sbjct 1 HSEGTFTSDYSKYMDNRRAKDFVQW#LMSTKRN 32 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 |||||||||||||||+||||||+||#||+| Sbjct 1 HSQGTFTSDYSKYLDNRRAQDFIQW#LMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRN 84 ||+|||++||||||++|||||||||#| |+||| Sbjct 1 HSEGTFSNDYSKYLETRRAQDFVQW#LKNSKRN 32 |
| Contains: Glucagon; Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 |||||||+|||||+|+|||||||||#||+| Sbjct 1 HSQGTFTNDYSKYMDTRRAQDFVQW#LMST 29 |
| Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 |||||||+|||||+|+|||||||||#||+| Sbjct 1 HSQGTFTNDYSKYMDTRRAQDFVQW#LMST 29 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 ||+||||||||||+|+|||+|||||#||+| Sbjct 1 HSEGTFTSDYSKYMDNRRAKDFVQW#LMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRN 84 ||+|||++||||||+ |+|||||+|#||| ||+ Sbjct 1 HSEGTFSNDYSKYLEDRKAQDFVRW#LMNNKRS 32 |
| Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 ||||+|||||||||||++|+||| |#|||| Sbjct 1 HSQGSFTSDYSKYLDSKQAKDFVIW#LMNT 29 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMN 80 ||+|||||||||||++++|+|||+|#||| Sbjct 1 HSEGTFTSDYSKYLENKQAKDFVRW#LMN 28 |
| deshis1, Desphe6, Glu9 | Query 54 SQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 |||| ||+||||||||||||||||#|||| Sbjct 1 SQGT-TSEYSKYLDSRRAQDFVQW#LMNT 27 |
| Monodelphis domestica | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGT 102 +|+++||| |||| +| |||| |#|++ | +| + ||+ || |+|| Sbjct 51 QRYAEGTFISDYSITMDKIMQQDFVNW#LLSQKGKKN--SWRHNITERKADGT 100 |
| Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||||||||||++++|+|||+|#||| || + + + |||+||||+|+ Sbjct 41 KRHSEGTFTSDYSKYLENKQAKDFVRW#LMNAKRGGSELQR------RHADGTFTNDM 91 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNR---NNIAKRHDEFERHAE 100 +||+ ||||+| + |||++ |+||| |#| + ++| + + | +|||| Sbjct 80 RRHADGTFTNDMTSYLDAKAARDFVSW#LARSDKSRRDGGDHLAENSEDKRHAE 132 |
| Xenopus laevis | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 | |||||||||||||||||||||||||||#||||||+ +++|+ ++|||||||||||| Sbjct 49 EVKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRS-GGLSRRNADYERHAEGTFTSDV 106 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 | +||++||+|+| ++||+ + + |++|#|+ | + + ||||||||+|+ Sbjct 138 EIERHAEGTYTNDVTEYLEEKATKAFIEW#LIKGK-------PKKIRYSRHAEGTFTNDM 189 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRH 92 ||++||||+| + ||+ + |++|| |#|+| + | ++ + | Sbjct 179 RHAEGTFTNDMTNYLEEKAAKEFVGW#LINGRPKRKDLLEEH 219 |
| Hoplobatrachus rugulosus | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 || |||||||||||||||||||||||||||#|||+||+ |+||+ +||||||||+|+|| Sbjct 48 NEVKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNSKRS-GGISKRNVQFERHAEGTYTNDV 106 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFE-RHAEGTFTSD 106 ||++|||||| + ||+ + |++|| |#|+ + || +| + |||+|+|||| Sbjct 134 RHAEGTFTSDMTSYLEEKAAKEFVDW#LIKGRPKRNFPDISAEEMDRRHADGSFTSD 189 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 +||++||+|+| +++|+ + |++|+ |#|+ | + ++ ||||||||||+ Sbjct 95 ERHAEGTYTNDVTQFLEEKAAKEFIDW#LIKGKPKKQRLS-------RHAEGTFTSDM 144 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 |+||+ |+||||++| || + ||+|+ |#++|| Sbjct 178 DRRHADGSFTSDFNKALDIKAAQEFLDW#IINT 209 |
| Bufo marinus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LM--NTKRNRNNIAKRHDEFERHAEGTFTSD 106 ||++|||||| + +|+ + |++|| |#|+ ||| ++++ | |||+|+|||| Sbjct 62 RHAEGTFTSDMTSFLEEKAAKEFVDW#LIKGRPKRNFSDVSAADDMDRRHADGSFTSD 118 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 +||++||+|+| +++|+ + |++|+ |#|+ | |+ ||||||||||+ Sbjct 23 ERHAEGTYTNDVTQFLEEKAAKEFIDW#LL------KGIPKK-QRLSRHAEGTFTSDM 72 Query 73 DFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 || ||#|||+||+ +++|+ +||||||||+|+|| Sbjct 1 DFAQW#LMNSKRS-GGMSRRNVQFERHAEGTYTNDV 34 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 |+||+ |+||||++| || + ||+|+ |#++|| Sbjct 107 DRRHADGSFTSDFNKALDIKAAQEFLDW#IINT 138 |
| Gallus gallus | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRN------------------RNNIA 89 || |||||||||||||||||||||||||||#||+|||| | |+ Sbjct 50 NEVKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMSTKRNGQQGQEDKENDKFPDQLSSNAIS 109 Query 90 KRHDEFERHAEGTFTSDV 107 ||| |||||||||+|||+ Sbjct 110 KRHSEFERHAEGTYTSDI 127 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAK---RHDEFERHAEGTFT 104 +| +||++||+||| + ||+ + |++|+ |#|+| + |+ | + |||+|||| Sbjct 113 SEFERHAEGTYTSDITSYLEGQAAKEFIAW#LVNGRGRRDFPEKALMAEEMGRRHADGTFT 172 Query 105 SDV 107 ||+ Sbjct 173 SDI 175 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNI 88 +||+ |||||| +| || |++|++|#|+||| + ++ Sbjct 164 RRHADGTFTSDINKILDDMAAKEFLKW#LINTKVTQRDL 201 |
| Meleagris gallopavo | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRN------------------RNNIA 89 || |||||||||||||||||||||||||||#||+|||| | |+ Sbjct 50 NEVKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMSTKRNGQQGQEDKENDEFPDQLSSNAIS 109 Query 90 KRHDEFERHAEGTFTSDV 107 ||| |||||||||+|||+ Sbjct 110 KRHSEFERHAEGTYTSDI 127 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAK---RHDEFERHAEGTFT 104 +| +||++||+||| + ||+ + |++|+ |#|+| + |+ | + |||+|||| Sbjct 113 SEFERHAEGTYTSDITSYLEGQAAKEFIAW#LVNGRGRRDFPEKALMAEEMGRRHADGTFT 172 Query 105 SDV 107 ||+ Sbjct 173 SDI 175 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNI 88 +||+ |||||| +| || |++|++|#|+||| + ++ Sbjct 164 RRHADGTFTSDINKILDDMAAKEFLKW#LINTKVTQRDL 201 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKR------------------NRNNIA 89 || |||||||||||||||||+|||||||||#|||||| + | +| Sbjct 48 NEVKRHSQGTFTSDYSKYLDTRRAQDFVQW#LMNTKRSGQQGVEEREKENLLDQLSSNGLA 107 Query 90 KRHDEFERHAEGTFTSDV 107 + | |+||||+| +|||+ Sbjct 108 RHHAEYERHADGRYTSDI 125 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAK---RHDEFERHAEGTFTS 105 | +||+ | +||| | ||+ + |++|+ |#|+| + |+ + + | |||+||||| Sbjct 112 EYERHADGRYTSDISSYLEGQAAKEFIAW#LVNGRGRRDFLEEAGTADDIGRRHADGTFTS 171 Query 106 D 106 | Query 106 D 106 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNI 88 +||+ |||||||++ || |+|++|#|+| | + ++ Sbjct 162 RRHADGTFTSDYNQLLDDIATQEFLKW#LINQKVTQRDL 199 |
| Capra hircus | Query 86 NNIAKRHDEFERHAEGTFTSDV 107 |||||||||||||||||||||| Query 86 NNIAKRHDEFERHAEGTFTSDV 107 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNR 85 +| +||++|||||| | ||+ + |++|+ |#|+ + | Sbjct 8 DEFERHAEGTFTSDVSSYLEGQAAKEFIAW#LVKGRGRR 45 |
| Oncorhynchus tshawytscha | Query 54 SQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 | |||++||||| + | |||||||#|||+||+ +|||+||+|||| Sbjct 2 SXGTFSNDYSKYQEERMAQDFVQW#LMNSKRS-------GAPSKRHADGTYTSDV 48 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNR 85 |||+ ||+||| | || + |+||| |#| + + | Sbjct 37 KRHADGTYTSDVSTYLQDQAAKDFVSW#LKSGRARR 71 |
| Ictalurus punctatus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 +|||+|||++||||||++|||||||||#|||+||+ |||+||+|||| Sbjct 49 RRHSEGTFSNDYSKYLETRRAQDFVQW#LMNSKRS-------DSPARRHADGTYTSDV 98 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEF--ERHAEGTFTSDV 107 +||+ ||+||| | || + |+||+ |#| + + + + | + || +|+||||| Sbjct 87 RRHADGTYTSDVSSYLQDQAAKDFITW#LKSGQPKPDAVETRSVDTLRRRHVDGSFTSDV 145 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRH 92 +|| |+|||| +| ||| |++++||#+||+ + + ||| Sbjct 134 RRHVDGSFTSDVNKVLDSLAAKEYLQW#VMNSPSSAS--GKRH 173 |
| Rattus norvegicus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHA 99 |+++||| |||| +| | |||| |#|+ | +|+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDFVNW#LLAQKGKKND--WKHNLTQREA 88 |
| Canis familiaris | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHA 99 |+++||| |||| +| | |||| |#|+ | +|+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDFVNW#LLAQKGKKND--WKHNITQREA 88 |
| Ambloplites rupestris | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSD 106 |||||||+||+ ||+ |+||||++|#||| | |+ |++ |||+|||| | Sbjct 1 HSQGTFTNDYTNYLEDRQAQDFIRW#LMNNK--RSGAAEK-----RHADGTFTDD 47 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDE--FERHAEGTFTSDV 107 +|||+ |||| | | + +||| #| + + | | | || ||+||||+ Sbjct 36 EKRHADGTFTDDASSDFYDQAIKDFVAK#LKSGQDRREPETDRRREAFSRRHVEGSFTSDI 95 |
| Pan troglodytes | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 ||||||||||||||||||||||||||||||#||||||| +++| ||||||||||| Sbjct 970 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRN-SSVA------YRHAEGTFTSDV 1022 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNR 85 ||++|||||| | ||+ + |++|+ |#|+ + | Sbjct 1012 RHAEGTFTSDVSSYLEGQAAKEFIAW#LVKGRGRR 1045 |
| Xenopus tropicalis | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 | ||||||||||||||||||||||||+||#||||||+ +++|+ ++|||||||||||| Sbjct 49 EVKRHSQGTFTSDYSKYLDSRRAQDFIQW#LMNTKRS-GGLSRRNADYERHAEGTFTSDV 106 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 +||++||+|+| ++||+ + |++|++|#|+| | + + ||||||||+|+ Sbjct 140 ERHAEGTYTNDVTEYLEEKAAKEFIEW#LINGK-------PKKIRYSRHAEGTFTNDM 189 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFE---RHAEGTFTSDV 107 ||++||||+| + ||+ + |++|| |#|+ | + | ++ | | |||+|+||+|+ Sbjct 179 RHAEGTFTNDMTNYLEEKAAKEFVGW#LIK-GRPKRNFSEAHSVEEMDRRHADGSFTNDI 236 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 |+||+ |+||+| +| || ||+|+ |#++||+ Sbjct 224 DRRHADGSFTNDINKVLDIIAAQEFLDW#VINTQ 256 |
| Danio rerio | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++||||||++|||||||||#||| ||| ++ +||||||+|||| Sbjct 48 KRHSEGTFSNDYSKYLETRRAQDFVQW#LMNAKRNGGSV-------KRHAEGTYTSDV 97 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#L 78 |||++||+||| | || + || || |#| Sbjct 86 KRHAEGTYTSDVSSYLQDQAAQSFVAW#L 113 |
| Bos taurus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSD 106 ||+ | ||||||+ | |+ +++ #|+ || |+|++ +||++ || + Sbjct 80 RHADGVFTSDYSRLLGQLSAKKYLES#LIG-KRVSNSISEDQGPIKRHSDAVFTDN 133 |
| Macaca mulatta | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSD 106 + + ||+ | ||||+|| | |+ +++ #|| || +||++ +||++ || + Sbjct 76 SRNARHADGVFTSDFSKLLGQLSAKKYLES#LMG-KRVSSNISEDPVPVKRHSDAVFTDN 133 |
| Heloderma suspectum | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKR------------------NRNNIA 89 || |||||||||||||||||+|||||||||#|||||| + | +| Sbjct 48 NEVKRHSQGTFTSDYSKYLDTRRAQDFVQW#LMNTKRSGQQGVEEREKENLLDQLSSNGLA 107 Query 90 KRHDEFERHAEGTFTSDV 107 + | |+||||+| +|||+ Sbjct 108 RHHAEYERHADGRYTSDI 125 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNR 85 | +||+ | +||| | ||+ + |++|+ |#|+| + | Sbjct 112 EYERHADGRYTSDISSYLEGQAAKEFIAW#LVNGRGRR 148 |
| Squalus acanthias | Query 67 DSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 |+|||+|||||#||+|||| |+ +||||||+|||| Sbjct 1 DNRRAKDFVQW#LMSTKRN-------GDKTKRHAEGTYTSDV 34 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#L--MNTKRNRNNIAKRHDEFERHAEGTF 103 ++ |||++||+||| | +|+ || #| | ++| +|+||| | || ||++ Sbjct 20 DKTKRHAEGTYTSDVDSLSDYFKAKRFVDS#LTSYNKRQNGRSISKRHVESTRHTEGSY 77 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRN---------NIAKRHDEFERHA 99 | ||++|++ | | ||+++ |+||+ |#|+ + | | + ||| Sbjct 68 ESTRHTEGSYR-DISSYLEAKAAKDFINW#LIKGRGRREFPEESKEIVNEVIPEEMDRRHA 126 Query 100 EGTFTSDV 107 +|||||++ Sbjct 127 DGTFTSEI 134 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFE 96 |+||+ |||||+ + ||+ |++|+ |#++|+| | | | | Sbjct 122 DRRHADGTFTSEINIVLDTIAAKEFLNW#ILNSK----TIQSRDSEIE 164 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRN 84 |+ ||||+| + |||++ |+||| |#| + ++ Sbjct 1 HADGTFTNDMTSYLDAKAARDFVSW#LARSDKS 32 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++||||||++|||||||||#| |+||| | |||+||+|||| Sbjct 50 KRHSEGTFSNDYSKYLETRRAQDFVQW#LKNSKRN--------GLFRRHADGTYTSDV 98 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#L 78 +||+ ||+||| | || + |+||| |#| Sbjct 87 RRHADGTYTSDVSSYLQDQAAKDFVSW#L 114 |
| Sebastes caurinus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++|||+||+ |+|||||+|#||| ||+ || +|||+||||||| Sbjct 48 KRHSEGTFSNDYSRYLEERKAQDFVRW#LMNNKRS------GADE-KRHADGTFTSDV 97 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDE--FERHAEGTFTSD 106 ++|||+ |||||| | || + +||| #| + + | + + | || +|+|||| Sbjct 84 DEKRHADGTFTSDVSSYLKDQAIKDFVNR#LKSGQVRRESETEWRGEAFSRRHVDGSFTSD 143 Query 107 V 107 | Query 107 V 107 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDE 94 +|| |+|||| +| ||| |++++ |#+| +| + + ++ |+ Sbjct 133 RRHVDGSFTSDVNKVLDSMAAKEYLLW#VMTSKPSGESKKRQEDQ 176 |
| Tetraodon nigroviridis | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++||||||++|||||||||#| |+||| + | |||+||+|||| Sbjct 787 KRHSEGTFSNDYSKYLETRRAQDFVQW#LKNSKRNGS-------LFRRHADGTYTSDV 836 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNR 85 +||+ ||+||| | || + |++|| |#| + | Sbjct 825 RRHADGTYTSDVSAYLQDQAAKEFVSW#LKTGRGRR 859 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++||||||++|||||||+|#|||+| | + +||||||+|||+ Sbjct 48 KRHSEGTFSNDYSKYLETRRAQDFVEW#LMNSKSNGGSA-------KRHAEGTYTSDI 97 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#L 78 |||++||+||| | +| + ||+|| |#| Sbjct 86 KRHAEGTYTSDISSFLRDQAAQNFVAW#L 113 |
| Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 || |+|||++ |||| + | |+||+ |#|||+||+ +|||+||+|||| Sbjct 50 KRQSEGTFSNYYSKYQEERMARDFLHW#LMNSKRS-------GAPSKRHADGTYTSDV 99 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHD--EFERHAEGTFTSDV 107 |||+ ||+||| | || + |+||| |#| + | + + + || +|+||||| Sbjct 88 KRHADGTYTSDVSTYLQDQAAKDFVSW#LKSGPARRESAEESMNGPMSRRHVDGSFTSDV 146 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHD 93 +|| |+|||| +| ||| |++++ |#+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSLAAKEYLLW#VMTSKTSGKSNKRQED 177 |
| Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++||||| + | |||||||#|||+||+ +|||+||+|||| Sbjct 50 KRHSEGTFSNDYSKYQEERMAQDFVQW#LMNSKRS-------GAPSKRHADGTYTSDV 99 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNI--AKRHDEFERHAEGTFTSDV 107 |||+ ||+||| | || + |+||| |#| + + | + ++ || +|+||||| Sbjct 88 KRHADGTYTSDVSTYLQDQAAKDFVSW#LKSGRARRESAEESRNGPMSRRHVDGSFTSDV 146 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHD 93 +|| |+|||| +| ||| |++++ |#+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSLAAKEYLLW#VMTSKTSGKSNKRQED 177 |
| Heloderma horridum | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMN 80 |||| |||||| || ++ + |++|#| | Sbjct 46 KRHSDGTFTSDLSKQMEEEAVRLFIEW#LKN 75 |
| Mus musculus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHA 99 |+++||| |||| +| | |||| |#|+ + +++ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDFVNW#LLAQRGKKSD--WKHNITQREA 88 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 |+ |+|||| +| ||+ |++|+ |#|++|| Sbjct 1 HADGSFTSDINKVLDTIAAKEFLNW#LISTK 30 |
| Protopterus dolloi | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 | |||||||| |||+| |+++ | | || #||+||| ++|||+| |||||||+|||+ Sbjct 28 EAKRHSQGTFMSDYAKLLEAKHALDLVQR#LMSTKRT-GAVSKRHNEIERHAEGTYTSDI 85 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFE-RHAEGTFTSD 106 || +||++||+||| | ||+ + +|++|#|| + |+ +| + |||+|| ||| Sbjct 71 NEIERHAEGTYTSDISSYLEGQAVNEFIRW#LMKGRGRRDFSETDIEEMDRRHADGTVTSD 130 Query 107 V 107 + Sbjct 131 I 131 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 |+||+ || ||| + |+| ++|+ |#|+| | Sbjct 119 DRRHADGTVTSDINSVLESIATREFLNW#LINAK 151 |
| Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSD 106 + + ||+ | ||||||+ | |+ +++ #|+ || ++|++ +||++ || + Sbjct 76 SRNARHADGVFTSDYSRLLGQISAKKYLES#LIG-KRISSSISEDPVPIKRHSDAVFTDN 133 |
| Agkistrodon piscivorus | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRH---DEFERHAEGTFTS 105 | +||+ ||+||| | ||+ + |++|+ |#|+| + |+ | | |||+||||| Sbjct 32 EYERHADGTYTSDISSYLEGQAAKEFIAW#LVNGRGRRDFSEGAHTGEDLGRRHADGTFTS 91 Query 106 D 106 | Query 106 D 106 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNI 88 +||+ |||||||+| || |+|++|#|+| | + ++ Sbjct 82 RRHADGTFTSDYNKLLDDIATQEFLKW#LINQKVTQRDL 119 Query 66 LDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 |+ | ++|+ #| + | +|+ | |+||||+||+|||+ Sbjct 9 LEDREKENFLDQ#LAS-----NGLARPHAEYERHADGTYTSDI 45 |
| Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 |||||+|||||||+|| ++|+||| |#|+|||| + + +||++|+||+|+ Sbjct 43 RHSQGSFTSDYSKHLDVKQAKDFVTW#LLNTKRRGVDAQAGANLEKRHSDGSFTNDM 98 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKR 83 | +|||| |+||+| + || |++|++|#| | Sbjct 84 NLEKRHSDGSFTNDMNVMLDRMSAKNFLEW#LKQQGR 119 |
| Arvicanthis ansorgei | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSD 106 + + ||+ | ||||||+ | |+ +++ #|+ || ++| + +||++ || + Sbjct 59 SRNARHADGVFTSDYSRLLGQISAKKYLES#LIG-KRISSSILEDPVPIKRHSDAVFTDN 116 |
| synthetic construct | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRN---NIAKRHDEFERHAEGTFT 104 +| +||++|||||| | ||+ + |++|+ |#|+ + |+ +| + |||+|+|+ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQAAKEFIAW#LVKGRGRRDFPEEVAIVEELGRRHADGSFS 152 Query 105 SDV 107 ++ Sbjct 153 DEM 155 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 +||+ |+|+ + + ||+ |+||+ |#|+ || Sbjct 144 RRHADGSFSDEMNTILDNLAARDFINW#LIQTK 175 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++||||||+ |+||+||+|#||| | |+ +|++ |||+||||||| Sbjct 51 KRHSEGTFSNDYSKYLEDRKAQEFVRW#LMNNK--RSGVAEK-----RHADGTFTSDV 100 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#L 78 +|||+ |||||| | || + +||| #| Sbjct 88 EKRHADGTFTSDVSSYLKDQAIKDFVDR#L 116 |
| Phodopus sungorus | Query 56 GTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRN---NIAKRHDEFERHAEGTFTSDV 107 |||||| | ||+ + |++|+ |#|+ + |+ + + |||+|+|+ ++ Sbjct 1 GTFTSDVSSYLEGQAAKEFIAW#LVKGRGRRDFPEEVTIVEELGRRHADGSFSDEM 55 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 +||+ |+|+ + + ||| +||+ |#|+ || Sbjct 44 RRHADGSFSDEMNTILDSLATRDFINW#LIQTK 75 |
| Neoceratodus forsteri | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 | |||||||| |||+| |++| | |||| #||||||| ++||| | |||||||+|||+ Sbjct 20 EAKRHSQGTFMSDYAKLLEARHALDFVQR#LMNTKRN-GGVSKRHSEIERHAEGTYTSDI 77 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEF-ERHAEGTFTSD 106 +| +||++||+||| | ||+ + | +||+|#|+ + |+ +| |||+|| |+| Sbjct 63 SEIERHAEGTYTSDISSYLEGQAANEFVRW#LLKGRGRRDFSDTDAEEMGRRHADGTITND 122 Query 107 V 107 + Sbjct 123 M 123 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 +||+ || |+| + |+| ++|+ |#|+|+| Sbjct 112 RRHADGTITNDMNNVLESIATREFLNW#LINSK 143 |
| Sus scrofa | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 |||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRN---NIAKRHDEFERHAEGTFT 104 +| +||++|||||| | ||+ + |++|+ |#|+ + |+ + + |||+|+|+ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQAAKEFIAW#LVKGRGRRDFPEEVTIVEELRRRHADGSFS 152 Query 105 SDV 107 ++ Sbjct 153 DEM 155 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 +||+ |+|+ + + ||+ +||+ |#|++|| Sbjct 144 RRHADGSFSDEMNTVLDNLATRDFINW#LLHTK 175 |
| Canis lupus familiaris | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRN---NIAKRHDEFERHAEGTFT 104 +| +||++|||||| | ||+ + |++|+ |#|+ + |+ +| + |||+|+|+ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQAAKEFIAW#LVKGRGRRDFPEEVAIVEEFRRRHADGSFS 152 Query 105 SDV 107 ++ Sbjct 153 DEM 155 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 +||+ |+|+ + + ||+ +||+ |#|+ || Sbjct 144 RRHADGSFSDEMNTVLDTLATRDFINW#LLQTK 175 |
| Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 |||||||+|||||||+|||||||||#||+|||+ | ||+||+|||| Sbjct 1 HSQGTFTNDYSKYLDTRRAQDFVQW#LMSTKRS-GGITXXXXXXXXHADGTYTSDV 54 Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNR 85 |+ ||+||| | || + |+ || |#| + | Sbjct 45 HADGTYTSDVSSYLQDQAAKKFVTW#LKQGQDRR 77 |
| Hydrolagus colliei | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 |||+ | |+||||||||+|| +|||||#|++|||| | |+ +|||+| +|||| Sbjct 30 KRHTDGIFSSDYSKYLDNRRTKDFVQW#LLSTKRNGANT----DKTKRHADGIYTSDV 82 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNN--IAKRHDEFERHAEGTFTS 105 ++ |||+ | +||| + | +++ ||+ #| | + +|+ ++||| | | | + + Sbjct 68 DKTKRHADGIYTSDVASLTDYLKSKRFVES#LSNYNKRQNDRRMSKRHVEGARRTEEDYPN 127 Query 106 D 106 | Query 106 D 106 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFE 96 | | ++ + +|+| ||+++ |+||+ |#|+ + | + |+ | | Sbjct 116 EGARRTEEDYPNDFSSYLEAKAAKDFIDW#LIK-GQGRRDFAEESREIE 162 |
| Heloderma suspectum, venom, Peptide, 39 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMN 80 | +|||||| || ++ + |++|#| | Sbjct 1 HGEGTFTSDLSKQMEEEAVRLFIEW#LKN 28 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | Query 56 GTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRN 86 ||+||| | || + |++|| |#| + |+ Sbjct 3 GTYTSDVSSYLQDQAAKEFVSW#LKTGRGRRD 33 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNR 85 |+ || ||| | +|+ + ++|+ |#|++ + | Sbjct 1 HADGTLTSDISSFLEKQATKEFIAW#LVSGRGRR 33 |
[Site 4] FTSDYSKYLD67-SRRAQDFVQW
Asp67  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe58 | Thr59 | Ser60 | Asp61 | Tyr62 | Ser63 | Lys64 | Tyr65 | Leu66 | Asp67 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser68 | Arg69 | Arg70 | Ala71 | Gln72 | Asp73 | Phe74 | Val75 | Gln76 | Trp77 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| ADPLSDPDQMNEDKRHSQGTFTSDYSKYLDSRRAQDFVQWLMNTKRNRNNIAKRHDEFER |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 129.00 | 9 | glucagon, isoform CRA_b |
| 2 | Pan troglodytes | 106.00 | 2 | PREDICTED: fibroblast activation protein, alpha su |
| 3 | N/A | 79.70 | 43 | 0 |
| 4 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | 64.70 | 1 | AAB31092.1 |
| 5 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | 61.60 | 1 | glucagon=proglucagon-derived 3.5 kda peptide |
| 6 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | 60.80 | 1 | glucagon II |
| 7 | Contains: Glucagon; Glucagon-like peptide | 60.10 | 4 | GLUC_AMICA Glucagon precursor |
| 8 | Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | 60.10 | 1 | glucagon |
| 9 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | 58.50 | 1 | glucagon G-29 |
| 10 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | 57.80 | 1 | GLUC1_ORENI Glucagon-1 (Glucagon I) gb |
| 11 | Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | 56.20 | 1 | GLUC_LAMFL Glucagon gb |
| 12 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | 53.50 | 1 | glucagon |
| 13 | deshis1, Desphe6, Glu9 | 53.50 | 1 | A Chain A, Nmr Solution Structure Of The Glucagon |
| 14 | Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | 47.00 | 1 | GLUC1_PETMA Glucagon-1 precursor |
| 15 | Gallus gallus | 44.30 | 4 | glucagon isoform 1 |
| 16 | Meleagris gallopavo | 44.30 | 2 | preproglucagon B |
| 17 | Agkistrodon piscivorus | 44.30 | 1 | proglucagon |
| 18 | Capra hircus | 43.90 | 1 | glucagon |
| 19 | Oncorhynchus tshawytscha | 43.50 | 1 | S78474_1 proglucagon |
| 20 | Monodelphis domestica | 42.70 | 3 | PREDICTED: similar to Gastric inhibitory polypepti |
| 21 | Xenopus laevis | 42.40 | 5 | proglucagon I |
| 22 | Rattus norvegicus | 42.40 | 3 | Glucose-dependent insulinotropic peptide |
| 23 | Xenopus tropicalis | 42.40 | 2 | glucagon |
| 24 | Canis familiaris | 42.40 | 2 | PREDICTED: similar to Gastric inhibitory polypepti |
| 25 | Danio rerio | 42.00 | 4 | hypothetical protein LOC494052 |
| 26 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | 42.00 | 1 | GLUC_HELSU Glucagon precursor |
| 27 | Heloderma suspectum | 41.60 | 2 | proglucagon |
| 28 | Hoplobatrachus rugulosus | 41.60 | 1 | AF324209_1 proglucagon |
| 29 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | 41.20 | 1 | glucagon-like peptide, GLP |
| 30 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | 41.20 | 1 | GLUC2_LOPAM Glucagon-2 precursor (Glucagon II) |
| 31 | Heloderma horridum | 40.80 | 1 | EXE3_HELHO Exendin-3 precursor emb |
| 32 | Bos taurus | 40.40 | 5 | PREDICTED: similar to Gastric inhibitory polypepti |
| 33 | Ictalurus punctatus | 40.40 | 3 | proglucagon I |
| 34 | Macaca mulatta | 40.40 | 3 | PREDICTED: gastric inhibitory polypeptide |
| 35 | Protopterus dolloi | 40.40 | 1 | proglucagon |
| 36 | Sebastes caurinus | 40.00 | 4 | proglucagon II |
| 37 | Tetraodon nigroviridis | 40.00 | 2 | unnamed protein product |
| 38 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | 40.00 | 2 | GLUC_CARAU Glucagon precursor |
| 39 | Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC2_ONCMY Glucagon-2 precursor (Glucagon II) |
| 40 | Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC1_ONCMY Glucagon-1 precursor (Glucagon I) |
| 41 | Neoceratodus forsteri | 39.70 | 1 | proglucagon |
| 42 | Mus musculus | 39.30 | 5 | gastric inhibitory polypeptide |
| 43 | Bufo marinus | 38.90 | 1 | proglucagon |
| 44 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | 38.90 | 1 | glucagon-like peptide-2, GLP-2=proglucagon-derived |
| 45 | Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | 38.10 | 1 | GLUC2_PETMA Glucagon-2 precursor |
| 46 | synthetic construct | 37.00 | 1 | Homo sapiens glucagon |
| 47 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | 36.60 | 1 | GLUC1_LOPAM Glucagon-1 precursor (Glucagon I) |
| 48 | Phodopus sungorus | 36.60 | 1 | glucagon precursor |
| 49 | Sus scrofa | 36.20 | 1 | glucagon |
| 50 | Canis lupus familiaris | 35.80 | 1 | glucagon |
| 51 | Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | 35.80 | 1 | GLUC_LEPSP Glucagon precursor |
| 52 | Heloderma suspectum, venom, Peptide, 39 aa | 35.40 | 1 | A Chain A, Solution Structure Of Exendin-4 In 30-V |
| 53 | Hydrolagus colliei | 35.40 | 1 | proglucagon |
| 54 | Squalus acanthias | 35.40 | 1 | proglucagon |
| 55 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | 34.70 | 1 | glucagon-like peptide-1, GLP-1=proglucagon-derived |
| 56 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | 34.70 | 1 | GLUC2_ORENI Glucagon-2 (Glucagon II) gb |
| 57 | Candida glabrata | 32.70 | 1 | unnamed protein product |
| 58 | Ambloplites rupestris | 32.30 | 1 | proglucagon |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 38 ADPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 38 ADPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 |
| Pan troglodytes | Query 38 ADPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDE 94 ||||||||||||||||||||||||||||||#||||||||||||||||| +++| || | Sbjct 960 ADPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN-SSVAYRHAE 1015 |
| N/A | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIA 89 |||||||||||||||#|||||||||||||||||+|||| Sbjct 1 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIA 37 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN 84 ||+||||||||||+|#+|||+|||||||+|||| Sbjct 1 HSEGTFTSDYSKYMD#NRRAKDFVQWLMSTKRN 32 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 |||||||||||||||#+||||||+||||+| Sbjct 1 HSQGTFTSDYSKYLD#NRRAQDFIQWLMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN 84 ||+|||++||||||+#+|||||||||| |+||| Sbjct 1 HSEGTFSNDYSKYLE#TRRAQDFVQWLKNSKRN 32 |
| Contains: Glucagon; Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 |||||||+|||||+|#+|||||||||||+| Sbjct 1 HSQGTFTNDYSKYMD#TRRAQDFVQWLMST 29 |
| Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 |||||||+|||||+|#+|||||||||||+| Sbjct 1 HSQGTFTNDYSKYMD#TRRAQDFVQWLMST 29 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 ||+||||||||||+|#+|||+|||||||+| Sbjct 1 HSEGTFTSDYSKYMD#NRRAKDFVQWLMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN 84 ||+|||++||||||+# |+|||||+|||| ||+ Sbjct 1 HSEGTFSNDYSKYLE#DRKAQDFVRWLMNNKRS 32 |
| Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 ||||+||||||||||#|++|+||| ||||| Sbjct 1 HSQGSFTSDYSKYLD#SKQAKDFVIWLMNT 29 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMN 80 ||+|||||||||||+#+++|+|||+|||| Sbjct 1 HSEGTFTSDYSKYLE#NKQAKDFVRWLMN 28 |
| deshis1, Desphe6, Glu9 | Query 54 SQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 |||| ||+||||||#|||||||||||||| Sbjct 1 SQGT-TSEYSKYLD#SRRAQDFVQWLMNT 27 |
| Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRH 92 ||||+|||||||||||+#+++|+|||+|||| || + + +|| Sbjct 41 KRHSEGTFTSDYSKYLE#NKQAKDFVRWLMNAKRGGSELQRRH 82 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNR---------NNIAKRHDE 94 +||+ ||||+| + |||#++ |+||| || + ++| |+ ||| | Sbjct 80 RRHADGTFTNDMTSYLD#AKAARDFVSWLARSDKSRRDGGDHLAENSEDKRHAE 132 |
| Gallus gallus | Query 38 ADPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN------------- 84 ++|| + |+|| |||||||||||||||||#||||||||||||+|||| Sbjct 40 SEPLDESRQLNEVKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMSTKRNGQQGQEDKENDKF 99 Query 85 -----RNNIAKRHDEFER 97 | |+||| |||| Sbjct 100 PDQLSSNAISKRHSEFER 117 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNI 88 +||+ |||||| +| ||# |++|++||+||| + ++ Sbjct 164 RRHADGTFTSDINKILD#DMAAKEFLKWLINTKVTQRDL 201 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRN 86 +| +||++||+||| + ||+# + |++|+ ||+| + |+ Sbjct 113 SEFERHAEGTYTSDITSYLE#GQAAKEFIAWLVNGRGRRD 151 |
| Meleagris gallopavo | Query 38 ADPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN------------- 84 ++|| + |+|| |||||||||||||||||#||||||||||||+|||| Sbjct 40 SEPLDESRQLNEVKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMSTKRNGQQGQEDKENDEF 99 Query 85 -----RNNIAKRHDEFER 97 | |+||| |||| Sbjct 100 PDQLSSNAISKRHSEFER 117 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNI 88 +||+ |||||| +| ||# |++|++||+||| + ++ Sbjct 164 RRHADGTFTSDINKILD#DMAAKEFLKWLINTKVTQRDL 201 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRN 86 +| +||++||+||| + ||+# + |++|+ ||+| + |+ Sbjct 113 SEFERHAEGTYTSDITSYLE#GQAAKEFIAWLVNGRGRRD 151 |
| Agkistrodon piscivorus | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNI 88 +||+ |||||||+| ||# |+|++||+| | + ++ Sbjct 82 RRHADGTFTSDYNKLLD#DIATQEFLKWLINQKVTQRDL 119 Query 49 EDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRN 86 | +||+ ||+||| | ||+# + |++|+ ||+| + |+ Sbjct 32 EYERHADGTYTSDISSYLE#GQAAKEFIAWLVNGRGRRD 69 |
| Capra hircus | Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNR 85 +| +||++|||||| | ||+# + |++|+ ||+ + | Sbjct 8 DEFERHAEGTFTSDVSSYLE#GQAAKEFIAWLVKGRGRR 45 |
| Oncorhynchus tshawytscha | Query 54 SQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRH 92 | |||++||||| +# | ||||||||||+||+ +||| Sbjct 2 SXGTFSNDYSKYQE#ERMAQDFVQWLMNSKRS-GAPSKRH 39 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNR 85 |||+ ||+||| | || # + |+||| || + + | Sbjct 37 KRHADGTYTSDVSTYLQ#DQAAKDFVSWLKSGRARR 71 |
| Monodelphis domestica | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 +|+++||| |||| +|# |||| ||++ | +| + ||+ || Sbjct 51 QRYAEGTFISDYSITMD#KIMQQDFVNWLLSQKGKKN--SWRHNITER 95 |
| Xenopus laevis | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 + + | +|+ | |||||||||||||||||#||||||||||||||||+ +++|+ ++|| Sbjct 39 EAVDDSEQLKEVKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRS-GELSRRNADYER 96 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 +||++|||||| ++ ||# + |++|+ ||+| ++ |++|+ |||| Sbjct 95 ERHAEGTFTSDVTQQLD#EKAAKEFIDWLINGGPSKEIISRRNAEFER 141 Query 50 DKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNI 88 |+||+ |+||+| +| ||# ||+|+ |++||+ ++ Sbjct 224 DRRHADGSFTNDINKVLD#IIAAQEFLDWVINTQETERDL 262 Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRN 86 ||++||||+| + ||+# + |++|| ||+ + || Sbjct 179 RHAEGTFTNDMTNYLE#EKAAKEFVGWLIKGRPKRN 213 Query 49 EDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDE 94 | +||++||+|+| ++||+# + |++|++||+ | + + || | Sbjct 138 EFERHAEGTYTNDVTEYLE#EKAAKEFIEWLIKGKPKKIRYS-RHAE 182 |
| Rattus norvegicus | Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNN 87 |+++||| |||| +|# | |||| ||+ | +|+ Sbjct 43 RYAEGTFISDYSIAMD#KIRQQDFVNWLLAQKGKKND 78 |
| Xenopus tropicalis | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 + + | |+ | |||||||||||||||||#|||||||+||||||||+ +++|+ ++|| Sbjct 39 EAVDDSQQLKEVKRHSQGTFTSDYSKYLD#SRRAQDFIQWLMNTKRS-GGLSRRNADYER 96 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 +||++|||||| +++||# + |++|+ ||+| ++ |++|+ + || Sbjct 95 ERHAEGTFTSDVTQHLD#EKAAKEFIDWLINGGPSKEIISRRNADVER 141 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDE 94 +||++||+|+| ++||+# + |++|++||+| | + + || | Sbjct 140 ERHAEGTYTNDVTEYLE#EKAAKEFIEWLINGKPKKIRYS-RHAE 182 Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRN 86 ||++||||+| + ||+# + |++|| ||+ + || Sbjct 179 RHAEGTFTNDMTNYLE#EKAAKEFVGWLIKGRPKRN 213 Query 50 DKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 |+||+ |+||+| +| ||# ||+|+ |++||+ Sbjct 224 DRRHADGSFTNDINKVLD#IIAAQEFLDWVINTQ 256 |
| Canis familiaris | Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNN 87 |+++||| |||| +|# | |||| ||+ | +|+ Sbjct 43 RYAEGTFISDYSIAMD#KIRQQDFVNWLLAQKGKKND 78 |
| Danio rerio | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDE 94 ||||+|||++||||||+#+|||||||||||| ||| ++ ||| | Sbjct 48 KRHSEGTFSNDYSKYLE#TRRAQDFVQWLMNAKRNGGSV-KRHAE 90 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWL 78 |||++||+||| | || # + || || || Sbjct 86 KRHAEGTYTSDVSSYLQ#DQAAQSFVAWL 113 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | Query 38 ADPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKR-------------- 83 |+|| | |+|| |||||||||||||||||#+||||||||||||||| Sbjct 38 AEPLDDSRQLNEVKRHSQGTFTSDYSKYLD#TRRAQDFVQWLMNTKRSGQQGVEEREKENL 97 Query 84 ----NRNNIAKRHDEFER 97 + | +|+ | |+|| Sbjct 98 LDQLSSNGLARHHAEYER 115 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNI 88 +||+ |||||||++ ||# |+|++||+| | + ++ Sbjct 162 RRHADGTFTSDYNQLLD#DIATQEFLKWLINQKVTQRDL 199 Query 49 EDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRN 86 | +||+ | +||| | ||+# + |++|+ ||+| + |+ Sbjct 112 EYERHADGRYTSDISSYLE#GQAAKEFIAWLVNGRGRRD 149 |
| Heloderma suspectum | Query 38 ADPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKR-------------- 83 |+|| | |+|| |||||||||||||||||#+||||||||||||||| Sbjct 38 AEPLDDSRQLNEVKRHSQGTFTSDYSKYLD#TRRAQDFVQWLMNTKRSGQQGVEEREKENL 97 Query 84 ----NRNNIAKRHDEFER 97 + | +|+ | |+|| Sbjct 98 LDQLSSNGLARHHAEYER 115 Query 49 EDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNR 85 | +||+ | +||| | ||+# + |++|+ ||+| + | Sbjct 112 EYERHADGRYTSDISSYLE#GQAAKEFIAWLVNGRGRR 148 |
| Hoplobatrachus rugulosus | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 + | | | || |||||||||||||||||#|||||||||||||+||+ |+||+ +||| Sbjct 39 EALDDSDHHNEVKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNSKRS-GGISKRNVQFER 96 Query 50 DKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 |+||+ |+||||++| ||# + ||+|+ |++|| Sbjct 178 DRRHADGSFTSDFNKALD#IKAAQEFLDWIINT 209 Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRN 86 ||++|||||| + ||+# + |++|| ||+ + || Sbjct 134 RHAEGTFTSDMTSYLE#EKAAKEFVDWLIKGRPKRN 168 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDE 94 +||++||+|+| +++|+# + |++|+ ||+ | + ++ || | Sbjct 95 ERHAEGTYTNDVTQFLE#EKAAKEFIDWLIKGKPKKQRLS-RHAE 137 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN 84 |+ ||||+| + |||#++ |+||| || + ++ Sbjct 1 HADGTFTNDMTSYLD#AKAARDFVSWLARSDKS 32 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRH 92 |++| + ||||+|||++||||||+#+|||||||||| |+| || + +|| Sbjct 40 LTEPIEPLNMKRHSEGTFSNDYSKYLE#TRRAQDFVQWLKNSK--RNGLFRRH 89 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWL 78 +||+ ||+||| | || # + |+||| || Sbjct 87 RRHADGTYTSDVSSYLQ#DQAAKDFVSWL 114 |
| Heloderma horridum | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMN 80 | | ++ |||| |||||| || ++# + |++|| | Sbjct 34 DSASSESFASKIKRHSDGTFTSDLSKQME#EEAVRLFIEWLKN 75 |
| Bos taurus | Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNN 87 |+++||| |||| +|# | |||| ||+ | +++ Sbjct 81 RYAEGTFISDYSIAMD#KIRQQDFVNWLLAQKGKKSD 116 |
| Ictalurus punctatus | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRH 92 +|||+|||++||||||+#+||||||||||||+||+ ++ |+|| Sbjct 49 RRHSEGTFSNDYSKYLE#TRRAQDFVQWLMNSKRS-DSPARRH 89 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRH 92 +|| |+|||| +| ||#| |++++||+||+ + + ||| Sbjct 134 RRHVDGSFTSDVNKVLD#SLAAKEYLQWVMNSPSSAS--GKRH 173 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWL 78 +||+ ||+||| | || # + |+||+ || Sbjct 87 RRHADGTYTSDVSSYLQ#DQAAKDFITWL 114 |
| Macaca mulatta | Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNN 87 |+++||| |||| +|# |||| ||+ | +|+ Sbjct 51 RYAEGTFISDYSIAMD#KIHQQDFVNWLLAQKGKKND 86 |
| Protopterus dolloi | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 + + + + | |||||||| |||+| |+#++ | | || ||+||| ++|||+| || Sbjct 18 EAVDESREHTEAKRHSQGTFMSDYAKLLE#AKHALDLVQRLMSTKRT-GAVSKRHNEIER 75 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 || +||++||+||| | ||+# + +|++||| + |+ +| +| Sbjct 71 NEIERHAEGTYTSDISSYLE#GQAVNEFIRWLMKGRGRRDFSETDIEEMDR 120 Query 42 SDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 |+ | |+||+ || ||| + |+#| ++|+ ||+| | Sbjct 111 SETDIEEMDRRHADGTVTSDINSVLE#SIATREFLNWLINAK 151 |
| Sebastes caurinus | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRH 92 |++| ++ ||||+|||++||||||+#+|||||||||| |+||| ++ +|| Sbjct 40 LAEPIELPNMKRHSEGTFSNDYSKYLE#TRRAQDFVQWLKNSKRN-GSLFRRH 90 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNR 85 +||+ ||+||| | || # + |++|| || + | Sbjct 88 RRHADGTYTSDVSSYLQ#DQAAKEFVYWLKTGRGRR 122 |
| Tetraodon nigroviridis | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRH 92 |++| ++ ||||+|||++||||||+#+|||||||||| |+||| ++ +|| Sbjct 777 LTEPSDLSNMKRHSEGTFSNDYSKYLE#TRRAQDFVQWLKNSKRN-GSLFRRH 827 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNR 85 +||+ ||+||| | || # + |++|| || + | Sbjct 825 RRHADGTYTSDVSAYLQ#DQAAKEFVSWLKTGRGRR 859 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDE 94 ||||+|||++||||||+#+|||||||+||||+| | + |||| | Sbjct 48 KRHSEGTFSNDYSKYLE#TRRAQDFVEWLMNSKSNGGS-AKRHAE 90 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWL 78 |||++||+||| | +| # + ||+|| || Sbjct 86 KRHAEGTYTSDISSFLR#DQAAQNFVAWL 113 |
| Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRH 92 || |+|||++ |||| +# | |+||+ ||||+||+ +||| Sbjct 50 KRQSEGTFSNYYSKYQE#ERMARDFLHWLMNSKRS-GAPSKRH 90 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNN 87 |||+ ||+||| | || # + |+||| || + | + Sbjct 88 KRHADGTYTSDVSTYLQ#DQAAKDFVSWLKSGPARRES 124 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHD 93 +|| |+|||| +| ||#| |++++ |+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLD#SLAAKEYLLWVMTSKTSGKSNKRQED 177 |
| Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | Query 39 DPL-SDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRH 92 ||| | ++ ||||+|||++||||| +# | ||||||||||+||+ +||| Sbjct 37 DPLLEDLMGVSNVKRHSEGTFSNDYSKYQE#ERMAQDFVQWLMNSKRS-GAPSKRH 90 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNN 87 |||+ ||+||| | || # + |+||| || + + | + Sbjct 88 KRHADGTYTSDVSTYLQ#DQAAKDFVSWLKSGRARRES 124 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHD 93 +|| |+|||| +| ||#| |++++ |+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLD#SLAAKEYLLWVMTSKTSGKSNKRQED 177 |
| Neoceratodus forsteri | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 +|+ + | |||||||| |||+| |+#+| | |||| ||||||| ++||| | || Sbjct 10 EPVDASREHTEAKRHSQGTFMSDYAKLLE#ARHALDFVQRLMNTKRN-GGVSKRHSEIER 67 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRN 86 +| +||++||+||| | ||+# + | +||+||+ + |+ Sbjct 63 SEIERHAEGTYTSDISSYLE#GQAANEFVRWLLKGRGRRD 101 Query 42 SDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 || | +||+ || |+| + |+#| ++|+ ||+|+| Sbjct 103 SDTDAEEMGRRHADGTITNDMNNVLE#SIATREFLNWLINSK 143 |
| Mus musculus | Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNN 87 |+++||| |||| +|# | |||| ||+ + +++ Sbjct 43 RYAEGTFISDYSIAMD#KIRQQDFVNWLLAQRGKKSD 78 |
| Bufo marinus | Query 50 DKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 |+||+ |+||||++| ||# + ||+|+ |++|| Sbjct 107 DRRHADGSFTSDFNKALD#IKAAQEFLDWIINT 138 Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRN 86 ||++|||||| + +|+# + |++|| ||+ + || Sbjct 62 RHAEGTFTSDMTSFLE#EKAAKEFVDWLIKGRPKRN 96 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDE 94 +||++||+|+| +++|+# + |++|+ ||+ + ++ || | Sbjct 23 ERHAEGTYTNDVTQFLE#EKAAKEFIDWLLKGIPKKQRLS-RHAE 65 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 |+ |+|||| +| ||#+ |++|+ ||++|| Sbjct 1 HADGSFTSDINKVLD#TIAAKEFLNWLISTK 30 |
| Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN------RNNIAKRHDE 94 |||||+|||||||+||# ++|+||| ||+|||| |+ ||| + Sbjct 43 RHSQGSFTSDYSKHLD#VKQAKDFVTWLLNTKRRGVDAQAGANLEKRHSD 91 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKR 83 | +|||| |+||+| + ||# |++|++|| | Sbjct 84 NLEKRHSDGSFTNDMNVMLD#RMSAKNFLEWLKQQGR 119 |
| synthetic construct | Query 38 ADPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 38 ADPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 +||+ |+|+ + + ||#+ |+||+ ||+ || Sbjct 144 RRHADGSFSDEMNTILD#NLAARDFINWLIQTK 175 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | Query 43 DPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRH 92 +| +++ ||||+|||++||||||+# |+||+||+|||| ||+ ||| Sbjct 43 EPRELSNMKRHSEGTFSNDYSKYLE#DRKAQEFVRWLMNNKRS-GVAEKRH 91 Query 50 DKRHSQGTFTSDYSKYLD#SRRAQDFVQWL 78 +|||+ |||||| | || # + +||| | Sbjct 88 EKRHADGTFTSDVSSYLK#DQAIKDFVDRL 116 |
| Phodopus sungorus | Query 56 GTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRN 86 |||||| | ||+# + |++|+ ||+ + |+ Sbjct 1 GTFTSDVSSYLE#GQAAKEFIAWLVKGRGRRD 31 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 +||+ |+|+ + + ||#| +||+ ||+ || Sbjct 44 RRHADGSFSDEMNTILD#SLATRDFINWLIQTK 75 |
| Sus scrofa | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 ||| ||||| |||||||||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 39 DPLDDPDQMTEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 +||+ |+|+ + + ||#+ +||+ ||++|| Sbjct 144 RRHADGSFSDEMNTVLD#NLATRDFINWLLHTK 175 |
| Canis lupus familiaris | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 +||+| |||||||||||||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 39 EPLNDLDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 +||+ |+|+ + + ||#+ +||+ ||+ || Sbjct 144 RRHADGSFSDEMNTVLD#TLATRDFINWLLQTK 175 |
| Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN 84 |||||||+|||||||#+|||||||||||+|||+ Sbjct 1 HSQGTFTNDYSKYLD#TRRAQDFVQWLMSTKRS 32 Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNR 85 |+ ||+||| | || # + |+ || || + | Sbjct 45 HADGTYTSDVSSYLQ#DQAAKKFVTWLKQGQDRR 77 |
| Heloderma suspectum, venom, Peptide, 39 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMN 80 | +|||||| || ++# + |++|| | Sbjct 1 HGEGTFTSDLSKQME#EEAVRLFIEWLKN 28 |
| Hydrolagus colliei | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNI--AKRH 92 + +|+| + + |||+ | |+||||||||#+|| +||||||++|||| | ||| Sbjct 18 ESVSNPSTVTDVKRHTDGIFSSDYSKYLD#NRRTKDFVQWLLSTKRNGANTDKTKRH 73 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNN--IAKRHDEFER 97 ++ |||+ | +||| + |# +++ ||+ | | + +|+ ++||| | | Sbjct 68 DKTKRHADGIYTSDVASLTD#YLKSKRFVESLSNYNKRQNDRRMSKRHVEGAR 119 Query 49 EDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFE 96 | | ++ + +|+| ||+#++ |+||+ ||+ + | + |+ | | Sbjct 116 EGARRTEEDYPNDFSSYLE#AKAAKDFIDWLIK-GQGRRDFAEESREIE 162 |
| Squalus acanthias | Query 44 PDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFE 96 |++| |+||+ |||||+ + ||#+ |++|+ |++|+| | | | | Sbjct 118 PEEM--DRRHADGTFTSEINIVLD#TIAAKEFLNWILNSK----TIQSRDSEIE 164 Query 67 D#SRRAQDFVQWLMNTKRNRNNIAKRHDE 94 |#+|||+|||||||+|||| + ||| | Sbjct 1 D#NRRAKDFVQWLMSTKRNGDK-TKRHAE 27 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWL--MNTKRNRNNIAKRHDEFER 97 ++ |||++||+||| |# +|+ || | | ++| +|+||| | | Sbjct 20 DKTKRHAEGTYTSDVDSLSD#YFKAKRFVDSLTSYNKRQNGRSISKRHVESTR 71 Query 49 EDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNR 85 | ||++|++ | | ||+#++ |+||+ ||+ + | Sbjct 68 ESTRHTEGSYR-DISSYLE#AKAAKDFINWLIKGRGRR 103 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNR 85 |+ || ||| | +|+# + ++|+ ||++ + | Sbjct 1 HADGTLTSDISSFLE#KQATKEFIAWLVSGRGRR 33 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | Query 56 GTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRN 86 ||+||| | || # + |++|| || + |+ Sbjct 3 GTYTSDVSSYLQ#DQAAKEFVSWLKTGRGRRD 33 |
| Candida glabrata | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAK 90 |+| | ||+ | | | | + +|#+ || |+ + | +| +| Sbjct 351 LADGDDDEEDEEDSSSTSESSYDEEID#NEEKQDIVEEEEDADENDDNFSK 400 |
| Ambloplites rupestris | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRH 92 |||||||+||+ ||+# |+||||++|||| ||+ ||| Sbjct 1 HSQGTFTNDYTNYLE#DRQAQDFIRWLMNNKRS-GAAEKRH 39 Query 50 DKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMN--------TKRNRNNIAKRHDE 94 +|||+ |||| | | # + +||| | + | | | ++|| | Sbjct 36 EKRHADGTFTDDASSDFY#DQAIKDFVAKLKSGQDRREPETDRRREAFSRRHVE 88 |
[Site 5] RAQDFVQWLM79-NTKRNRNNIA
Met79  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg70 | Ala71 | Gln72 | Asp73 | Phe74 | Val75 | Gln76 | Trp77 | Leu78 | Met79 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn80 | Thr81 | Lys82 | Arg83 | Asn84 | Arg85 | Asn86 | Asn87 | Ile88 | Ala89 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DKRHSQGTFTSDYSKYLDSRRAQDFVQWLMNTKRNRNNIAKRHDEFERHAEGTFTSDVSS |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 123.00 | 10 | glucagon, isoform CRA_b |
| 2 | N/A | 79.70 | 45 | 0 |
| 3 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | 64.70 | 1 | AAB31092.1 |
| 4 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | 61.60 | 1 | glucagon=proglucagon-derived 3.5 kda peptide |
| 5 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | 60.80 | 1 | glucagon II |
| 6 | Contains: Glucagon; Glucagon-like peptide | 60.10 | 4 | GLUC_AMICA Glucagon precursor |
| 7 | Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | 60.10 | 1 | glucagon |
| 8 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | 58.50 | 1 | glucagon G-29 |
| 9 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | 57.80 | 1 | GLUC1_ORENI Glucagon-1 (Glucagon I) gb |
| 10 | Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | 56.20 | 1 | GLUC_LAMFL Glucagon gb |
| 11 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | 53.50 | 1 | glucagon |
| 12 | deshis1, Desphe6, Glu9 | 53.50 | 1 | A Chain A, Nmr Solution Structure Of The Glucagon |
| 13 | Monodelphis domestica | 48.90 | 3 | PREDICTED: similar to Gastric inhibitory polypepti |
| 14 | Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | 48.90 | 1 | GLUC1_PETMA Glucagon-1 precursor |
| 15 | Xenopus laevis | 47.00 | 5 | proglucagon II |
| 16 | Hoplobatrachus rugulosus | 46.60 | 1 | AF324209_1 proglucagon |
| 17 | Bufo marinus | 46.60 | 1 | proglucagon |
| 18 | Gallus gallus | 46.20 | 4 | glucagon isoform 1 |
| 19 | Meleagris gallopavo | 46.20 | 2 | preproglucagon B |
| 20 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | 44.70 | 1 | GLUC_HELSU Glucagon precursor |
| 21 | Oncorhynchus tshawytscha | 43.50 | 1 | S78474_1 proglucagon |
| 22 | Ictalurus punctatus | 43.10 | 3 | proglucagon I |
| 23 | Ambloplites rupestris | 43.10 | 1 | proglucagon |
| 24 | Rattus norvegicus | 42.70 | 6 | Glucose-dependent insulinotropic peptide |
| 25 | Canis familiaris | 42.70 | 2 | PREDICTED: similar to Gastric inhibitory polypepti |
| 26 | Capra hircus | 42.70 | 1 | glucagon |
| 27 | Pan troglodytes | 42.40 | 2 | PREDICTED: fibroblast activation protein, alpha su |
| 28 | Xenopus tropicalis | 42.40 | 2 | glucagon |
| 29 | Danio rerio | 42.00 | 6 | hypothetical protein LOC494052 |
| 30 | Bos taurus | 42.00 | 5 | vasoactive intestinal peptide |
| 31 | Squalus acanthias | 41.60 | 1 | proglucagon |
| 32 | Agkistrodon piscivorus | 41.60 | 1 | proglucagon |
| 33 | Macaca mulatta | 41.20 | 3 | PREDICTED: vasoactive intestinal peptide |
| 34 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | 41.20 | 1 | glucagon-like peptide, GLP |
| 35 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | 41.20 | 1 | GLUC2_LOPAM Glucagon-2 precursor (Glucagon II) |
| 36 | Sebastes caurinus | 40.80 | 4 | proglucagon I |
| 37 | Heloderma suspectum | 40.80 | 2 | proglucagon |
| 38 | Tetraodon nigroviridis | 40.00 | 3 | unnamed protein product |
| 39 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | 40.00 | 2 | GLUC_CARAU Glucagon precursor |
| 40 | Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC2_ONCMY Glucagon-2 precursor (Glucagon II) |
| 41 | Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC1_ONCMY Glucagon-1 precursor (Glucagon I) |
| 42 | Heloderma horridum | 40.00 | 1 | EXE3_HELHO Exendin-3 precursor emb |
| 43 | Mus musculus | 39.70 | 6 | Gastric inhibitory polypeptide |
| 44 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | 38.90 | 1 | glucagon-like peptide-2, GLP-2=proglucagon-derived |
| 45 | Protopterus dolloi | 38.90 | 1 | proglucagon |
| 46 | Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | 38.10 | 1 | VIP_MOUSE VIP peptides precursor |
| 47 | Arvicanthis ansorgei | 37.40 | 1 | vasoactive intestinal polypeptide |
| 48 | synthetic construct | 37.00 | 1 | Homo sapiens glucagon |
| 49 | Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | 37.00 | 1 | GLUC2_PETMA Glucagon-2 precursor |
| 50 | Phodopus sungorus | 36.60 | 1 | glucagon precursor |
| 51 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | 36.60 | 1 | GLUC1_LOPAM Glucagon-1 precursor (Glucagon I) |
| 52 | Sus scrofa | 36.20 | 1 | glucagon |
| 53 | Neoceratodus forsteri | 36.20 | 1 | proglucagon |
| 54 | Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | 35.80 | 1 | GLUC_LEPSP Glucagon precursor |
| 55 | Canis lupus familiaris | 35.80 | 1 | glucagon |
| 56 | Heloderma suspectum, venom, Peptide, 39 aa | 35.40 | 1 | A Chain A, Solution Structure Of Exendin-4 In 30-V |
| 57 | Hydrolagus colliei | 35.00 | 1 | proglucagon |
| 58 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | 34.70 | 1 | GLUC2_ORENI Glucagon-2 (Glucagon II) gb |
| 59 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | 34.70 | 1 | glucagon-like peptide-1, GLP-1=proglucagon-derived |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSD 106 ||||||||||||||||||||||||||||||#||||||||||||||||||||||||||| Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSD 106 |
| N/A | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIA 89 |||||||||||||||||||||||||||#|||||+|||| Sbjct 1 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIA 37 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRN 84 ||+||||||||||+|+|||+|||||||#+|||| Sbjct 1 HSEGTFTSDYSKYMDNRRAKDFVQWLM#STKRN 32 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 |||||||||||||||+||||||+||||#+| Sbjct 1 HSQGTFTSDYSKYLDNRRAQDFIQWLM#ST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRN 84 ||+|||++||||||++|||||||||| #|+||| Sbjct 1 HSEGTFSNDYSKYLETRRAQDFVQWLK#NSKRN 32 |
| Contains: Glucagon; Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 |||||||+|||||+|+|||||||||||#+| Sbjct 1 HSQGTFTNDYSKYMDTRRAQDFVQWLM#ST 29 |
| Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 |||||||+|||||+|+|||||||||||#+| Sbjct 1 HSQGTFTNDYSKYMDTRRAQDFVQWLM#ST 29 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 ||+||||||||||+|+|||+|||||||#+| Sbjct 1 HSEGTFTSDYSKYMDNRRAKDFVQWLM#ST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRN 84 ||+|||++||||||+ |+|||||+|||#| ||+ Sbjct 1 HSEGTFSNDYSKYLEDRKAQDFVRWLM#NNKRS 32 |
| Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 ||||+|||||||||||++|+||| |||#|| Sbjct 1 HSQGSFTSDYSKYLDSKQAKDFVIWLM#NT 29 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#N 80 ||+|||||||||||++++|+|||+|||#| Sbjct 1 HSEGTFTSDYSKYLENKQAKDFVRWLM#N 28 |
| deshis1, Desphe6, Glu9 | Query 54 SQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 |||| ||+||||||||||||||||||#|| Sbjct 1 SQGT-TSEYSKYLDSRRAQDFVQWLM#NT 27 |
| Monodelphis domestica | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGT 102 +|+++||| |||| +| |||| ||+#+ | +| + ||+ || |+|| Sbjct 51 QRYAEGTFISDYSITMDKIMQQDFVNWLL#SQKGKKN--SWRHNITERKADGT 100 |
| Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||||||||||++++|+|||+|||#| || + + + |||+||||+|++| Sbjct 41 KRHSEGTFTSDYSKYLENKQAKDFVRWLM#NAKRGGSELQR------RHADGTFTNDMTS 93 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNR---NNIAKRHDEFERHAE 100 +||+ ||||+| + |||++ |+||| || # + ++| + + | +|||| Sbjct 80 RRHADGTFTNDMTSYLDAKAARDFVSWLA#RSDKSRRDGGDHLAENSEDKRHAE 132 |
| Xenopus laevis | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVS 108 |||||||||||||||||||||||||||||#||||+ +++|+ ++||||||||||||+ Sbjct 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRS-GGLSRRNADYERHAEGTFTSDVT 107 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 +||++||+|+| ++||+ + + |++||+# | + + ||||||||+|+++ Sbjct 140 ERHAEGTYTNDVTEYLEEKATKAFIEWLI#KGK-------PKKIRYSRHAEGTFTNDMTN 191 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRH 92 ||++||||+| + ||+ + |++|| ||+#| + | ++ + | Sbjct 179 RHAEGTFTNDMTNYLEEKAAKEFVGWLI#NGRPKRKDLLEEH 219 |
| Hoplobatrachus rugulosus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVS 108 |||||||||||||||||||||||||||||#|+||+ |+||+ +||||||||+|+||+ Sbjct 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NSKRS-GGISKRNVQFERHAEGTYTNDVT 107 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 +||++||+|+| +++|+ + |++|+ ||+# | + ++ ||||||||||++| Sbjct 95 ERHAEGTYTNDVTQFLEEKAAKEFIDWLI#KGKPKKQRLS-------RHAEGTFTSDMTS 146 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFE-RHAEGTFTSD 106 ||++|||||| + ||+ + |++|| ||+# + || +| + |||+|+|||| Sbjct 134 RHAEGTFTSDMTSYLEEKAAKEFVDWLI#KGRPKRNFPDISAEEMDRRHADGSFTSD 189 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 |+||+ |+||||++| || + ||+|+ |++#|| Sbjct 178 DRRHADGSFTSDFNKALDIKAAQEFLDWII#NT 209 |
| Bufo marinus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM--#NTKRNRNNIAKRHDEFERHAEGTFTSD 106 ||++|||||| + +|+ + |++|| ||+ # ||| ++++ | |||+|+|||| Sbjct 62 RHAEGTFTSDMTSFLEEKAAKEFVDWLIKG#RPKRNFSDVSAADDMDRRHADGSFTSD 118 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 +||++||+|+| +++|+ + |++|+ ||+# | |+ ||||||||||++| Sbjct 23 ERHAEGTYTNDVTQFLEEKAAKEFIDWLL#------KGIPKK-QRLSRHAEGTFTSDMTS 74 Query 73 DFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVS 108 || ||||#|+||+ +++|+ +||||||||+|+||+ Sbjct 1 DFAQWLM#NSKRS-GGMSRRNVQFERHAEGTYTNDVT 35 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 |+||+ |+||||++| || + ||+|+ |++#|| Sbjct 107 DRRHADGSFTSDFNKALDIKAAQEFLDWII#NT 138 |
| Gallus gallus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRN------------------RNNIAKRH 92 |||||||||||||||||||||||||||||#+|||| | |+||| Sbjct 53 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#STKRNGQQGQEDKENDKFPDQLSSNAISKRH 112 Query 93 DEFERHAEGTFTSDVSS 109 |||||||||+|||++| Sbjct 113 SEFERHAEGTYTSDITS 129 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAK---RHDEFERHAEGTFTSDV 107 +||++||+||| + ||+ + |++|+ ||+#| + |+ | + |||+||||||+ Sbjct 116 ERHAEGTYTSDITSYLEGQAAKEFIAWLV#NGRGRRDFPEKALMAEEMGRRHADGTFTSDI 175 Query 108 S 108 + Sbjct 176 N 176 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNI 88 +||+ |||||| +| || |++|++||+#||| + ++ Sbjct 164 RRHADGTFTSDINKILDDMAAKEFLKWLI#NTKVTQRDL 201 |
| Meleagris gallopavo | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRN------------------RNNIAKRH 92 |||||||||||||||||||||||||||||#+|||| | |+||| Sbjct 53 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#STKRNGQQGQEDKENDEFPDQLSSNAISKRH 112 Query 93 DEFERHAEGTFTSDVSS 109 |||||||||+|||++| Sbjct 113 SEFERHAEGTYTSDITS 129 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAK---RHDEFERHAEGTFTSDV 107 +||++||+||| + ||+ + |++|+ ||+#| + |+ | + |||+||||||+ Sbjct 116 ERHAEGTYTSDITSYLEGQAAKEFIAWLV#NGRGRRDFPEKALMAEEMGRRHADGTFTSDI 175 Query 108 S 108 + Sbjct 176 N 176 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNI 88 +||+ |||||| +| || |++|++||+#||| + ++ Sbjct 164 RRHADGTFTSDINKILDDMAAKEFLKWLI#NTKVTQRDL 201 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKR------------------NRNNIAKRH 92 |||||||||||||||||+|||||||||||#|||| + | +|+ | Sbjct 51 KRHSQGTFTSDYSKYLDTRRAQDFVQWLM#NTKRSGQQGVEEREKENLLDQLSSNGLARHH 110 Query 93 DEFERHAEGTFTSDVSS 109 |+||||+| +|||+|| Sbjct 111 AEYERHADGRYTSDISS 127 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAK---RHDEFERHAEGTFTSD 106 +||+ | +||| | ||+ + |++|+ ||+#| + |+ + + | |||+|||||| Sbjct 114 ERHADGRYTSDISSYLEGQAAKEFIAWLV#NGRGRRDFLEEAGTADDIGRRHADGTFTSD 172 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNI 88 +||+ |||||||++ || |+|++||+#| | + ++ Sbjct 162 RRHADGTFTSDYNQLLDDIATQEFLKWLI#NQKVTQRDL 199 |
| Oncorhynchus tshawytscha | Query 54 SQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 | |||++||||| + | |||||||||#|+||+ +|||+||+|||||+ Sbjct 2 SXGTFSNDYSKYQEERMAQDFVQWLM#NSKRS-------GAPSKRHADGTYTSDVST 50 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNR 85 |||+ ||+||| | || + |+||| || #+ + | Sbjct 37 KRHADGTYTSDVSTYLQDQAAKDFVSWLK#SGRARR 71 |
| Ictalurus punctatus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 +|||+|||++||||||++|||||||||||#|+||+ |||+||+|||||| Sbjct 49 RRHSEGTFSNDYSKYLETRRAQDFVQWLM#NSKRS-------DSPARRHADGTYTSDVSS 100 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEF--ERHAEGTFTSDVS 108 +||+ ||+||| | || + |+||+ || #+ + + + | + || +|+|||||+ Sbjct 87 RRHADGTYTSDVSSYLQDQAAKDFITWLK#SGQPKPDAVETRSVDTLRRRHVDGSFTSDVN 146 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRH 92 +|| |+|||| +| ||| |++++||+|#|+ + + ||| Sbjct 134 RRHVDGSFTSDVNKVLDSLAAKEYLQWVM#NSPSSAS--GKRH 173 |
| Ambloplites rupestris | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 |||||||+||+ ||+ |+||||++|||#| | |+ |++ |||+|||| | || Sbjct 1 HSQGTFTNDYTNYLEDRQAQDFIRWLM#NNK--RSGAAEK-----RHADGTFTDDASS 50 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDE--FERHAEGTFTSDV 107 +|||+ |||| | | + +||| | #+ + | | | || ||+||||+ Sbjct 36 EKRHADGTFTDDASSDFYDQAIKDFVAKLK#SGQDRREPETDRRREAFSRRHVEGSFTSDI 95 Query 108 S 108 + Sbjct 96 N 96 |
| Rattus norvegicus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHA 99 |+++||| |||| +| | |||| ||+# | +|+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDFVNWLL#AQKGKKND--WKHNLTQREA 88 |
| Canis familiaris | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHA 99 |+++||| |||| +| | |||| ||+# | +|+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDFVNWLL#AQKGKKND--WKHNITQREA 88 |
| Capra hircus | Query 86 NNIAKRHDEFERHAEGTFTSDVSS 109 |||||||||||||||||||||||| Query 86 NNIAKRHDEFERHAEGTFTSDVSS 109 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNR 85 +||++|||||| | ||+ + |++|+ ||+# + | Sbjct 11 ERHAEGTFTSDVSSYLEGQAAKEFIAWLV#KGRGRR 45 |
| Pan troglodytes | Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 ||||||||||||||||||||||||||||||#||||| +++| ||||||||||||| Sbjct 972 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRN-SSVA------YRHAEGTFTSDVSS 1024 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNR 85 ||++|||||| | ||+ + |++|+ ||+# + | Sbjct 1012 RHAEGTFTSDVSSYLEGQAAKEFIAWLV#KGRGRR 1045 |
| Xenopus tropicalis | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVS 108 ||||||||||||||||||||||||+||||#||||+ +++|+ ++||||||||||||+ Sbjct 51 KRHSQGTFTSDYSKYLDSRRAQDFIQWLM#NTKRS-GGLSRRNADYERHAEGTFTSDVT 107 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 +||++||+|+| ++||+ + |++|++||+#| | + + ||||||||+|+++ Sbjct 140 ERHAEGTYTNDVTEYLEEKAAKEFIEWLI#NGK-------PKKIRYSRHAEGTFTNDMTN 191 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFE---RHAEGTFTSDVS 108 ||++||||+| + ||+ + |++|| ||+# | + | ++ | | |||+|+||+|++ Sbjct 179 RHAEGTFTNDMTNYLEEKAAKEFVGWLI#K-GRPKRNFSEAHSVEEMDRRHADGSFTNDIN 237 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 |+||+ |+||+| +| || ||+|+ |++#||+ Sbjct 224 DRRHADGSFTNDINKVLDIIAAQEFLDWVI#NTQ 256 |
| Danio rerio | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++||||||++|||||||||||#| ||| ++ +||||||+|||||| Sbjct 48 KRHSEGTFSNDYSKYLETRRAQDFVQWLM#NAKRNGGSV-------KRHAEGTYTSDVSS 99 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWL 78 |||++||+||| | || + || || || Sbjct 86 KRHAEGTYTSDVSSYLQDQAAQSFVAWL 113 |
| Bos taurus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSD 106 ||+ | ||||||+ | |+ +++ |+# || |+|++ +||++ || + Sbjct 80 RHADGVFTSDYSRLLGQLSAKKYLESLI#G-KRVSNSISEDQGPIKRHSDAVFTDN 133 |
| Squalus acanthias | Query 67 DSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 |+|||+|||||||#+|||| |+ +||||||+|||| | Sbjct 1 DNRRAKDFVQWLM#STKRN-------GDKTKRHAEGTYTSDVDS 36 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWL--M#NTKRNRNNIAKRHDEFERHAEGTFTSDVS 108 |||++||+||| | +|+ || | #| ++| +|+||| | || ||++ |+| Sbjct 23 KRHAEGTYTSDVDSLSDYFKAKRFVDSLTSY#NKRQNGRSISKRHVESTRHTEGSY-RDIS 81 Query 109 S 109 | Query 109 S 109 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRN---------NIAKRHDEFERHAEGT 102 ||++|++ | | ||+++ |+||+ ||+# + | | + |||+|| Sbjct 71 RHTEGSYR-DISSYLEAKAAKDFINWLI#KGRGRREFPEESKEIVNEVIPEEMDRRHADGT 129 Query 103 FTSDVS 108 |||+++ Sbjct 130 FTSEIN 135 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFE 96 |+||+ |||||+ + ||+ |++|+ |++#|+| | | | | Sbjct 122 DRRHADGTFTSEINIVLDTIAAKEFLNWIL#NSK----TIQSRDSEIE 164 |
| Agkistrodon piscivorus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRH---DEFERHAEGTFTSD 106 +||+ ||+||| | ||+ + |++|+ ||+#| + |+ | | |||+|||||| Sbjct 34 ERHADGTYTSDISSYLEGQAAKEFIAWLV#NGRGRRDFSEGAHTGEDLGRRHADGTFTSD 92 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNI 88 +||+ |||||||+| || |+|++||+#| | + ++ Sbjct 82 RRHADGTFTSDYNKLLDDIATQEFLKWLI#NQKVTQRDL 119 Query 66 LDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 |+ | ++|+ | #+ | +|+ | |+||||+||+|||+|| Sbjct 9 LEDREKENFLDQLA#S-----NGLARPHAEYERHADGTYTSDISS 47 |
| Macaca mulatta | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSD 106 ||+ | ||||+|| | |+ +++ ||# || +||++ +||++ || + Sbjct 80 RHADGVFTSDFSKLLGQLSAKKYLESLM#G-KRVSSNISEDPVPVKRHSDAVFTDN 133 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRN 84 |+ ||||+| + |||++ |+||| || # + ++ Sbjct 1 HADGTFTNDMTSYLDAKAARDFVSWLA#RSDKS 32 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++||||||++|||||||||| #|+||| | |||+||+|||||| Sbjct 50 KRHSEGTFSNDYSKYLETRRAQDFVQWLK#NSKRN--------GLFRRHADGTYTSDVSS 100 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWL 78 +||+ ||+||| | || + |+||| || Sbjct 87 RRHADGTYTSDVSSYLQDQAAKDFVSWL 114 |
| Sebastes caurinus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++|||+||+ |+|||||+|||#| ||+ || +|||+||||||||| Sbjct 48 KRHSEGTFSNDYSRYLEERKAQDFVRWLM#NNKRS------GADE-KRHADGTFTSDVSS 99 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDE--FERHAEGTFTSDV 107 +|||+ |||||| | || + +||| | #+ + | + + | || +|+||||| Sbjct 85 EKRHADGTFTSDVSSYLKDQAIKDFVNRLK#SGQVRRESETEWRGEAFSRRHVDGSFTSDV 144 Query 108 S 108 + Sbjct 145 N 145 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDE 94 +|| |+|||| +| ||| |++++ |+|# +| + + ++ |+ Sbjct 133 RRHVDGSFTSDVNKVLDSMAAKEYLLWVM#TSKPSGESKKRQEDQ 176 |
| Heloderma suspectum | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKR------------------NRNNIAKRH 92 |||||||||||||||||+|||||||||||#|||| + | +|+ | Sbjct 51 KRHSQGTFTSDYSKYLDTRRAQDFVQWLM#NTKRSGQQGVEEREKENLLDQLSSNGLARHH 110 Query 93 DEFERHAEGTFTSDVSS 109 |+||||+| +|||+|| Sbjct 111 AEYERHADGRYTSDISS 127 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNR 85 +||+ | +||| | ||+ + |++|+ ||+#| + | Sbjct 114 ERHADGRYTSDISSYLEGQAAKEFIAWLV#NGRGRR 148 |
| Tetraodon nigroviridis | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++||||||++|||||||||| #|+||| + | |||+||+|||||+ Sbjct 787 KRHSEGTFSNDYSKYLETRRAQDFVQWLK#NSKRNGS-------LFRRHADGTYTSDVSA 838 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNR 85 +||+ ||+||| | || + |++|| || # + | Sbjct 825 RRHADGTYTSDVSAYLQDQAAKEFVSWLK#TGRGRR 859 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++||||||++|||||||+|||#|+| | + +||||||+|||+|| Sbjct 48 KRHSEGTFSNDYSKYLETRRAQDFVEWLM#NSKSNGGSA-------KRHAEGTYTSDISS 99 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWL 78 |||++||+||| | +| + ||+|| || Sbjct 86 KRHAEGTYTSDISSFLRDQAAQNFVAWL 113 |
| Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 || |+|||++ |||| + | |+||+ |||#|+||+ +|||+||+|||||+ Sbjct 50 KRQSEGTFSNYYSKYQEERMARDFLHWLM#NSKRS-------GAPSKRHADGTYTSDVST 101 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHD--EFERHAEGTFTSDVS 108 |||+ ||+||| | || + |+||| || #+ | + + + || +|+|||||+ Sbjct 88 KRHADGTYTSDVSTYLQDQAAKDFVSWLK#SGPARRESAEESMNGPMSRRHVDGSFTSDVN 147 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHD 93 +|| |+|||| +| ||| |++++ |+|# +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSLAAKEYLLWVM#TSKTSGKSNKRQED 177 |
| Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++||||| + | |||||||||#|+||+ +|||+||+|||||+ Sbjct 50 KRHSEGTFSNDYSKYQEERMAQDFVQWLM#NSKRS-------GAPSKRHADGTYTSDVST 101 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNI--AKRHDEFERHAEGTFTSDVS 108 |||+ ||+||| | || + |+||| || #+ + | + ++ || +|+|||||+ Sbjct 88 KRHADGTYTSDVSTYLQDQAAKDFVSWLK#SGRARRESAEESRNGPMSRRHVDGSFTSDVN 147 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHD 93 +|| |+|||| +| ||| |++++ |+|# +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSLAAKEYLLWVM#TSKTSGKSNKRQED 177 |
| Heloderma horridum | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#N 80 |||| |||||| || ++ + |++|| #| Sbjct 46 KRHSDGTFTSDLSKQMEEEAVRLFIEWLK#N 75 |
| Mus musculus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHA 99 |+++||| |||| +| | |||| ||+# + +++ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDFVNWLL#AQRGKKSD--WKHNITQREA 88 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 |+ |+|||| +| ||+ |++|+ ||+#+|| Sbjct 1 HADGSFTSDINKVLDTIAAKEFLNWLI#STK 30 |
| Protopterus dolloi | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 |||||||| |||+| |+++ | | || ||#+||| ++|||+| |||||||+|||+|| Sbjct 30 KRHSQGTFMSDYAKLLEAKHALDLVQRLM#STKRT-GAVSKRHNEIERHAEGTYTSDISS 87 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFE-RHAEGTFTSDVSS 109 +||++||+||| | ||+ + +|++|||# + |+ +| + |||+|| |||++| Sbjct 74 ERHAEGTYTSDISSYLEGQAVNEFIRWLM#KGRGRRDFSETDIEEMDRRHADGTVTSDINS 133 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 |+||+ || ||| + |+| ++|+ ||+#| | Sbjct 119 DRRHADGTVTSDINSVLESIATREFLNWLI#NAK 151 |
| Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSD 106 ||+ | ||||||+ | |+ +++ |+# || ++|++ +||++ || + Sbjct 80 RHADGVFTSDYSRLLGQISAKKYLESLI#G-KRISSSISEDPVPIKRHSDAVFTDN 133 |
| Arvicanthis ansorgei | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSD 106 ||+ | ||||||+ | |+ +++ |+# || ++| + +||++ || + Sbjct 63 RHADGVFTSDYSRLLGQISAKKYLESLI#G-KRISSSILEDPVPIKRHSDAVFTDN 116 |
| synthetic construct | Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRN---NIAKRHDEFERHAEGTFTSDV 107 +||++|||||| | ||+ + |++|+ ||+# + |+ +| + |||+|+|+ ++ Sbjct 96 ERHAEGTFTSDVSSYLEGQAAKEFIAWLV#KGRGRRDFPEEVAIVEELGRRHADGSFSDEM 155 Query 108 SS 109 ++ Sbjct 156 NT 157 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 +||+ |+|+ + + ||+ |+||+ ||+# || Sbjct 144 RRHADGSFSDEMNTILDNLAARDFINWLI#QTK 175 |
| Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVS 108 |||||+|||||||+|| ++|+||| ||+#|||| + + +||++|+||+|++ Sbjct 43 RHSQGSFTSDYSKHLDVKQAKDFVTWLL#NTKRRGVDAQAGANLEKRHSDGSFTNDMN 99 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKR 83 +|||| |+||+| + || |++|++|| # | Sbjct 86 EKRHSDGSFTNDMNVMLDRMSAKNFLEWLK#QQGR 119 |
| Phodopus sungorus | Query 56 GTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRN---NIAKRHDEFERHAEGTFTSDVSS 109 |||||| | ||+ + |++|+ ||+# + |+ + + |||+|+|+ ++++ Sbjct 1 GTFTSDVSSYLEGQAAKEFIAWLV#KGRGRRDFPEEVTIVEELGRRHADGSFSDEMNT 57 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 +||+ |+|+ + + ||| +||+ ||+# || Sbjct 44 RRHADGSFSDEMNTILDSLATRDFINWLI#QTK 75 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++||||||+ |+||+||+|||#| | |+ +|++ |||+||||||||| Sbjct 51 KRHSEGTFSNDYSKYLEDRKAQEFVRWLM#NNK--RSGVAEK-----RHADGTFTSDVSS 102 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWL 78 +|||+ |||||| | || + +||| | Sbjct 88 EKRHADGTFTSDVSSYLKDQAIKDFVDRL 116 |
| Sus scrofa | Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 ||||||||||||||||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRN---NIAKRHDEFERHAEGTFTSDV 107 +||++|||||| | ||+ + |++|+ ||+# + |+ + + |||+|+|+ ++ Sbjct 96 ERHAEGTFTSDVSSYLEGQAAKEFIAWLV#KGRGRRDFPEEVTIVEELRRRHADGSFSDEM 155 Query 108 SS 109 ++ Sbjct 156 NT 157 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 +||+ |+|+ + + ||+ +||+ ||+#+|| Sbjct 144 RRHADGSFSDEMNTVLDNLATRDFINWLL#HTK 175 |
| Neoceratodus forsteri | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 |||||||| |||+| |++| | |||| ||#||||| ++||| | |||||||+|||+|| Sbjct 22 KRHSQGTFMSDYAKLLEARHALDFVQRLM#NTKRN-GGVSKRHSEIERHAEGTYTSDISS 79 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEF-ERHAEGTFTSDVSS 109 +||++||+||| | ||+ + | +||+||+# + |+ +| |||+|| |+|+++ Sbjct 66 ERHAEGTYTSDISSYLEGQAANEFVRWLL#KGRGRRDFSDTDAEEMGRRHADGTITNDMNN 125 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 +||+ || |+| + |+| ++|+ ||+#|+| Sbjct 112 RRHADGTITNDMNNVLESIATREFLNWLI#NSK 143 |
| Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 |||||||+|||||||+|||||||||||#+|||+ | ||+||+|||||| Sbjct 1 HSQGTFTNDYSKYLDTRRAQDFVQWLM#STKRS-GGITXXXXXXXXHADGTYTSDVSS 56 Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNR 85 |+ ||+||| | || + |+ || || # + | Sbjct 45 HADGTYTSDVSSYLQDQAAKKFVTWLK#QGQDRR 77 |
| Canis lupus familiaris | Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 ||||||||||||||||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRN---NIAKRHDEFERHAEGTFTSDV 107 +||++|||||| | ||+ + |++|+ ||+# + |+ +| + |||+|+|+ ++ Sbjct 96 ERHAEGTFTSDVSSYLEGQAAKEFIAWLV#KGRGRRDFPEEVAIVEEFRRRHADGSFSDEM 155 Query 108 SS 109 ++ Sbjct 156 NT 157 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 +||+ |+|+ + + ||+ +||+ ||+# || Sbjct 144 RRHADGSFSDEMNTVLDTLATRDFINWLL#QTK 175 |
| Heloderma suspectum, venom, Peptide, 39 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#N 80 | +|||||| || ++ + |++|| #| Sbjct 1 HGEGTFTSDLSKQMEEEAVRLFIEWLK#N 28 |
| Hydrolagus colliei | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 |||+ | |+||||||||+|| +||||||+#+|||| | |+ +|||+| +||||+| Sbjct 30 KRHTDGIFSSDYSKYLDNRRTKDFVQWLL#STKRNGANT----DKTKRHADGIYTSDVAS 84 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNN--IAKRHDEFERHAEGTFTSDVS 108 |||+ | +||| + | +++ ||+ | #| + +|+ ++||| | | | + +| | Sbjct 71 KRHADGIYTSDVASLTDYLKSKRFVESLS#NYNKRQNDRRMSKRHVEGARRTEEDYPNDFS 130 Query 109 S 109 | Query 109 S 109 Query 51 KRHSQGT------FTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFE 96 ||| +| + +|+| ||+++ |+||+ ||+# + | + |+ | | Sbjct 112 KRHVEGARRTEEDYPNDFSSYLEAKAAKDFIDWLI#K-GQGRRDFAEESREIE 162 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | Query 56 GTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRN 86 ||+||| | || + |++|| || # + |+ Sbjct 3 GTYTSDVSSYLQDQAAKEFVSWLK#TGRGRRD 33 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNR 85 |+ || ||| | +|+ + ++|+ ||+#+ + | Sbjct 1 HADGTLTSDISSFLEKQATKEFIAWLV#SGRGRR 33 |
* References
[PubMed ID: 18996102] Ruano G, Bernene J, Windemuth A, Bower B, Wencker D, Seip RL, Kocherla M, Holford TR, Petit WA, Hanks S, Physiogenomic comparison of edema and BMI in patients receiving rosiglitazone or pioglitazone. Clin Chim Acta. 2009 Feb;400(1-2):48-55. Epub 2008 Oct 26.
[PubMed ID: 19077438] ... Chung WK, Patki A, Matsuoka N, Boyer BB, Liu N, Musani SK, Goropashnaya AV, Tan PL, Katsanis N, Johnson SB, Gregersen PK, Allison DB, Leibel RL, Tiwari HK, Analysis of 30 genes (355 SNPS) related to energy homeostasis for association with adiposity in European-American and Yup'ik Eskimo populations. Hum Hered. 2009;67(3):193-205. Epub 2008 Dec 15.
[PubMed ID: 18832087] ... Liu X, Murali SG, Holst JJ, Ney DM, Enteral nutrients potentiate the intestinotrophic action of glucagon-like peptide-2 in association with increased insulin-like growth factor-I responses in rats. Am J Physiol Regul Integr Comp Physiol. 2008 Dec;295(6):R1794-802. Epub
[PubMed ID: 18760331] ... Qin Z, Sun Z, Huang J, Hu Y, Wu Z, Mei B, Mutated recombinant human glucagon-like peptide-1 protects SH-SY5Y cells from apoptosis induced by amyloid-beta peptide (1-42). Neurosci Lett. 2008 Oct 31;444(3):217-21. Epub 2008 Aug 22.
[PubMed ID: 18298441] ... Hellstrom PM, Naslund E, Edholm T, Schmidt PT, Kristensen J, Theodorsson E, Holst JJ, Efendic S, GLP-1 suppresses gastrointestinal motility and inhibits the migrating motor complex in healthy subjects and patients with irritable bowel syndrome. Neurogastroenterol Motil. 2008 Jun;20(6):649-59. Epub 2008 Feb 19.
[PubMed ID: 11278902] ... Bertenshaw GP, Turk BE, Hubbard SJ, Matters GL, Bylander JE, Crisman JM, Cantley LC, Bond JS, Marked differences between metalloproteases meprin A and B in substrate and peptide bond specificity. J Biol Chem. 2001 Apr 20;276(16):13248-55. Epub 2001 Jan 22.
[PubMed ID: 2753890] ... Orskov C, Bersani M, Johnsen AH, Hojrup P, Holst JJ, Complete sequences of glucagon-like peptide-1 from human and pig small intestine. J Biol Chem. 1989 Aug 5;264(22):12826-9.
[PubMed ID: 2901414] ... Drucker DJ, Asa S, Glucagon gene expression in vertebrate brain. J Biol Chem. 1988 Sep 25;263(27):13475-8.
[PubMed ID: 3569278] ... Novak U, Wilks A, Buell G, McEwen S, Identical mRNA for preproglucagon in pancreas and gut. Eur J Biochem. 1987 May 4;164(3):553-8.
[PubMed ID: 3725587] ... White JW, Saunders GF, Structure of the human glucagon gene. Nucleic Acids Res. 1986 Jun 25;14(12):4719-30.
[PubMed ID: 6546710] ... Schroeder WT, Lopez LC, Harper ME, Saunders GF, Localization of the human glucagon gene (GCG) to chromosome segment 2q36----37. Cytogenet Cell Genet. 1984;38(1):76-9.
[PubMed ID: 7340337] ... Kargel HJ, Dettmer R, Etzold G, Kirschke H, Bohley P, Langner J, Action of rat liver cathepsin L on glucagon. Acta Biol Med Ger. 1981;40(9):1139-43.