XSB2232 : PREDICTED: similar to proglucagon [Equus caballus]
[ CaMP Format ]
This entry is computationally expanded from SB0054
* Basic Information
| Organism | Equus caballus (horse) |
| Protein Names | similar to proglucagon |
| Gene Names | LOC100051551; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 1 mRNA, 2 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| XP_001494318 | XM_001494268 | 100051551 | N/A | N/A | N/A | N/A | N/A |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 179 aa
Average Mass: 20.951 kDa
Monoisotopic Mass: 20.938 kDa
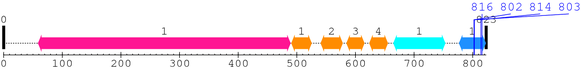
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Hormone_2 1. | 53 | 80 | 66.4 | 1.1e-16 |
| --- cleavage 74 (inside Hormone_2 53..80) --- | ||||
| --- cleavage 69 (inside Hormone_2 53..80) --- | ||||
| --- cleavage 77 (inside Hormone_2 53..80) --- | ||||
| --- cleavage 67 (inside Hormone_2 53..80) --- | ||||
| --- cleavage 79 (inside Hormone_2 53..80) --- | ||||
| Hormone_2 2. | 98 | 125 | 52.6 | 1.5e-12 |
| Hormone_2 3. | 146 | 173 | 46.6 | 9.8e-11 |
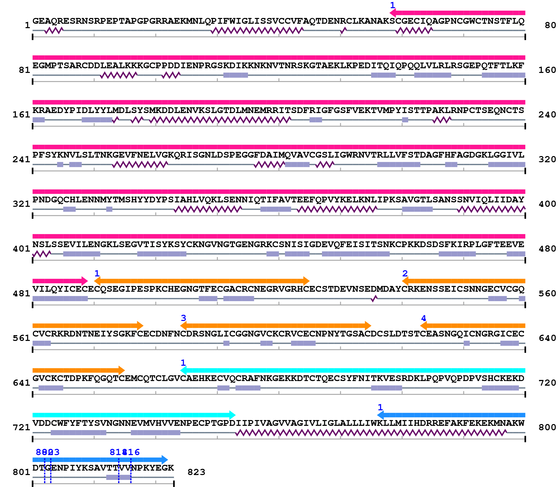
3. Sequence Information
Fasta Sequence: XSB2232.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
5 [sites]
Cleavage sites (±10aa)
[Site 1] YLDSRRAQDF74-VQWLMNTKRN
Phe74  Val
Val
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Tyr65 | Leu66 | Asp67 | Ser68 | Arg69 | Arg70 | Ala71 | Gln72 | Asp73 | Phe74 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Val75 | Gln76 | Trp77 | Leu78 | Met79 | Asn80 | Thr81 | Lys82 | Arg83 | Asn84 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DQMNEDKRHSQGTFTSDYSKYLDSRRAQDFVQWLMNTKRNKNNIAKRHDEFERHAEGTFT |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 128.00 | 10 | glucagon, isoform CRA_b |
| 2 | N/A | 80.90 | 44 | 0 |
| 3 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | 64.70 | 1 | AAB31092.1 |
| 4 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | 61.60 | 1 | glucagon=proglucagon-derived 3.5 kda peptide |
| 5 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | 60.80 | 1 | glucagon II |
| 6 | Contains: Glucagon; Glucagon-like peptide | 60.10 | 4 | GLUC_AMICA Glucagon precursor |
| 7 | Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | 60.10 | 1 | glucagon |
| 8 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | 58.50 | 1 | glucagon G-29 |
| 9 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | 57.80 | 1 | GLUC1_ORENI Glucagon-1 (Glucagon I) gb |
| 10 | Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | 56.20 | 1 | GLUC_LAMFL Glucagon gb |
| 11 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | 53.50 | 1 | glucagon |
| 12 | deshis1, Desphe6, Glu9 | 53.50 | 1 | A Chain A, Nmr Solution Structure Of The Glucagon |
| 13 | Monodelphis domestica | 50.10 | 3 | PREDICTED: similar to Gastric inhibitory polypepti |
| 14 | Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | 48.50 | 1 | GLUC1_PETMA Glucagon-1 precursor |
| 15 | Hoplobatrachus rugulosus | 46.60 | 1 | AF324209_1 proglucagon |
| 16 | Gallus gallus | 46.20 | 4 | glucagon isoform 1 |
| 17 | Meleagris gallopavo | 46.20 | 2 | preproglucagon B |
| 18 | Xenopus laevis | 45.80 | 5 | proglucagon II |
| 19 | Bufo marinus | 44.70 | 1 | proglucagon |
| 20 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | 44.70 | 1 | GLUC_HELSU Glucagon precursor |
| 21 | Rattus norvegicus | 43.90 | 5 | Glucose-dependent insulinotropic peptide |
| 22 | Canis familiaris | 43.90 | 2 | PREDICTED: similar to Gastric inhibitory polypepti |
| 23 | Ictalurus punctatus | 43.10 | 3 | proglucagon I |
| 24 | Xenopus tropicalis | 42.70 | 2 | glucagon |
| 25 | Capra hircus | 42.70 | 1 | glucagon |
| 26 | Bos taurus | 42.40 | 5 | vasoactive intestinal peptide |
| 27 | Oncorhynchus tshawytscha | 42.40 | 1 | S78474_1 proglucagon |
| 28 | Danio rerio | 42.00 | 6 | hypothetical protein LOC494052 |
| 29 | Macaca mulatta | 42.00 | 3 | PREDICTED: vasoactive intestinal peptide |
| 30 | Pan troglodytes | 42.00 | 2 | PREDICTED: gastric inhibitory polypeptide |
| 31 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | 41.20 | 1 | glucagon-like peptide, GLP |
| 32 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | 41.20 | 1 | GLUC2_LOPAM Glucagon-2 precursor (Glucagon II) |
| 33 | Mus musculus | 40.80 | 6 | gastric inhibitory polypeptide |
| 34 | Sebastes caurinus | 40.80 | 4 | proglucagon I |
| 35 | Heloderma suspectum | 40.40 | 2 | proglucagon |
| 36 | Squalus acanthias | 40.40 | 1 | proglucagon |
| 37 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | 40.00 | 2 | GLUC_CARAU Glucagon precursor |
| 38 | Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC2_ONCMY Glucagon-2 precursor (Glucagon II) |
| 39 | Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC1_ONCMY Glucagon-1 precursor (Glucagon I) |
| 40 | Heloderma horridum | 40.00 | 1 | EXE3_HELHO Exendin-3 precursor emb |
| 41 | Tetraodon nigroviridis | 39.30 | 3 | unnamed protein product |
| 42 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | 38.90 | 1 | glucagon-like peptide-2, GLP-2=proglucagon-derived |
| 43 | Protopterus dolloi | 38.90 | 1 | proglucagon |
| 44 | Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | 38.90 | 1 | VIP_MOUSE VIP peptides precursor |
| 45 | Ambloplites rupestris | 38.10 | 1 | proglucagon |
| 46 | Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | 38.10 | 1 | GLUC2_PETMA Glucagon-2 precursor |
| 47 | Arvicanthis ansorgei | 38.10 | 1 | vasoactive intestinal polypeptide |
| 48 | synthetic construct | 37.00 | 1 | Homo sapiens glucagon |
| 49 | Phodopus sungorus | 36.60 | 1 | glucagon precursor |
| 50 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | 36.60 | 1 | GLUC1_LOPAM Glucagon-1 precursor (Glucagon I) |
| 51 | Sus scrofa | 36.20 | 1 | glucagon |
| 52 | Neoceratodus forsteri | 36.20 | 1 | proglucagon |
| 53 | Canis lupus familiaris | 35.80 | 1 | glucagon |
| 54 | Heloderma suspectum, venom, Peptide, 39 aa | 35.40 | 1 | A Chain A, Solution Structure Of Exendin-4 In 30-V |
| 55 | Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | 35.40 | 1 | GLUC_LEPSP Glucagon precursor |
| 56 | Hydrolagus colliei | 34.30 | 1 | proglucagon |
| 57 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | 33.50 | 1 | glucagon-like peptide-1, GLP-1=proglucagon-derived |
| 58 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | 33.50 | 1 | GLUC2_ORENI Glucagon-2 (Glucagon II) gb |
| 59 | Agkistrodon piscivorus | 33.50 | 1 | proglucagon |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ||||||||||||||||||||||||||||||#||||||||||+||||||||||||||||||| Sbjct 128 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 187 |
| N/A | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIA 89 ||||||||||||||||||||||#||||||||||||||| Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIA 89 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRN 84 ||+||||||||||+|+|||+||#|||||+|||| Sbjct 1 HSEGTFTSDYSKYMDNRRAKDF#VQWLMSTKRN 32 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 |||||||||||||||+||||||#+||||+| Sbjct 1 HSQGTFTSDYSKYLDNRRAQDF#IQWLMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRN 84 ||+|||++||||||++||||||#|||| |+||| Sbjct 1 HSEGTFSNDYSKYLETRRAQDF#VQWLKNSKRN 32 |
| Contains: Glucagon; Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 |||||||+|||||+|+||||||#|||||+| Sbjct 1 HSQGTFTNDYSKYMDTRRAQDF#VQWLMST 29 |
| Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 |||||||+|||||+|+||||||#|||||+| Sbjct 1 HSQGTFTNDYSKYMDTRRAQDF#VQWLMST 29 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 ||+||||||||||+|+|||+||#|||||+| Sbjct 1 HSEGTFTSDYSKYMDNRRAKDF#VQWLMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRN 84 ||+|||++||||||+ |+||||#|+|||| ||+ Sbjct 1 HSEGTFSNDYSKYLEDRKAQDF#VRWLMNNKRS 32 |
| Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 ||||+|||||||||||++|+||#| ||||| Sbjct 1 HSQGSFTSDYSKYLDSKQAKDF#VIWLMNT 29 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMN 80 ||+|||||||||||++++|+||#|+|||| Sbjct 1 HSEGTFTSDYSKYLENKQAKDF#VRWLMN 28 |
| deshis1, Desphe6, Glu9 | Query 54 SQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 |||| ||+|||||||||||||#||||||| Sbjct 1 SQGT-TSEYSKYLDSRRAQDF#VQWLMNT 27 |
| Monodelphis domestica | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGT 102 +|+++||| |||| +| |||#| ||++ | || + ||+ || |+|| Sbjct 51 QRYAEGTFISDYSITMDKIMQQDF#VNWLLSQKGKKN--SWRHNITERKADGT 100 |
| Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ||||+|||||||||||++++|+||#|+|||| || + + + |||+|||| Sbjct 41 KRHSEGTFTSDYSKYLENKQAKDF#VRWLMNAKRGGSELQR------RHADGTFT 88 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLM---NTKRNKNNIAKRHDEFERHAE 100 +||+ ||||+| + |||++ |+||#| || ++|+ + + | +|||| Sbjct 80 RRHADGTFTNDMTSYLDAKAARDF#VSWLARSDKSRRDGGDHLAENSEDKRHAE 132 |
| Hoplobatrachus rugulosus | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 | || ||||||||||||||||||||||||#||||||+||+ |+||+ +||||||||+| Sbjct 45 DHHNEVKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNSKRS-GGISKRNVQFERHAEGTYT 103 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 +||++||+|+| +++|+ + |++|#+ ||+ | | ++ |||||||| Sbjct 95 ERHAEGTYTNDVTQFLEEKAAKEF#IDWLIKGKPKKQRLS-------RHAEGTFT 141 Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLM--NTKRNKNNIAKRHDEFERHAEGTFT 104 ||++|||||| + ||+ + |++|#| ||+ ||| +|+ + |||+|+|| Sbjct 134 RHAEGTFTSDMTSYLEEKAAKEF#VDWLIKGRPKRNFPDISAEEMD-RRHADGSFT 187 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 |+||+ |+||||++| || + ||+|#+ |++|| Sbjct 178 DRRHADGSFTSDFNKALDIKAAQEF#LDWIINT 209 |
| Gallus gallus | Query 46 QMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRN------------------KNN 87 |+|| ||||||||||||||||||||||||#|||||+|||| | Sbjct 48 QLNEVKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMSTKRNGQQGQEDKENDKFPDQLSSNA 107 Query 88 IAKRHDEFERHAEGTFT 104 |+||| |||||||||+| Sbjct 108 ISKRHSEFERHAEGTYT 124 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAK---RHDEFERHAEGTFT 104 +| +||++||+||| + ||+ + |++|#+ ||+| + ++ | + |||+|||| Sbjct 113 SEFERHAEGTYTSDITSYLEGQAAKEF#IAWLVNGRGRRDFPEKALMAEEMGRRHADGTFT 172 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNI 88 +||+ |||||| +| || |++|#++||+||| + ++ Sbjct 164 RRHADGTFTSDINKILDDMAAKEF#LKWLINTKVTQRDL 201 |
| Meleagris gallopavo | Query 46 QMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRN------------------KNN 87 |+|| ||||||||||||||||||||||||#|||||+|||| | Sbjct 48 QLNEVKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMSTKRNGQQGQEDKENDEFPDQLSSNA 107 Query 88 IAKRHDEFERHAEGTFT 104 |+||| |||||||||+| Sbjct 108 ISKRHSEFERHAEGTYT 124 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAK---RHDEFERHAEGTFT 104 +| +||++||+||| + ||+ + |++|#+ ||+| + ++ | + |||+|||| Sbjct 113 SEFERHAEGTYTSDITSYLEGQAAKEF#IAWLVNGRGRRDFPEKALMAEEMGRRHADGTFT 172 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNI 88 +||+ |||||| +| || |++|#++||+||| + ++ Sbjct 164 RRHADGTFTSDINKILDDMAAKEF#LKWLINTKVTQRDL 201 |
| Xenopus laevis | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 +|+ | ||||||||||||||||||||||||#|||||||||+ +++|+ ++||||||||| Sbjct 45 EQLKEVKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRS-GGLSRRNADYERHAEGTFT 103 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 | +||++||+|+| ++||+ + + |#++||+ | | + |||||||| Sbjct 138 EIERHAEGTYTNDVTEYLEEKATKAF#IEWLIKGKPKK-------IRYSRHAEGTFT 186 Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRH 92 ||++||||+| + ||+ + |++|#| ||+| + + ++ + | Sbjct 179 RHAEGTFTNDMTNYLEEKAAKEF#VGWLINGRPKRKDLLEEH 219 |
| Bufo marinus | Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLM--NTKRNKNNIAKRHDEFERHAEGTFT 104 ||++|||||| + +|+ + |++|#| ||+ ||| ++++ | |||+|+|| Sbjct 62 RHAEGTFTSDMTSFLEEKAAKEF#VDWLIKGRPKRNFSDVSAADDMDRRHADGSFT 116 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 +||++||+|+| +++|+ + |++|#+ ||+ | ++ |||||||| Sbjct 23 ERHAEGTYTNDVTQFLEEKAAKEF#IDWLLKGIPKKQRLS-------RHAEGTFT 69 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNT 81 |+||+ |+||||++| || + ||+|#+ |++|| Sbjct 107 DRRHADGSFTSDFNKALDIKAAQEF#LDWIINT 138 Query 73 DF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ||# |||||+||+ +++|+ +||||||||+| Sbjct 1 DF#AQWLMNSKRS-GGMSRRNVQFERHAEGTYT 31 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | Query 46 QMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKR------------------NKNN 87 |+|| |||||||||||||||||+||||||#||||||||| + | Sbjct 46 QLNEVKRHSQGTFTSDYSKYLDTRRAQDF#VQWLMNTKRSGQQGVEEREKENLLDQLSSNG 105 Query 88 IAKRHDEFERHAEGTFT 104 +|+ | |+||||+| +| Sbjct 106 LARHHAEYERHADGRYT 122 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAK---RHDEFERHAEGTFT 104 | +||+ | +||| | ||+ + |++|#+ ||+| + ++ + + | |||+|||| Sbjct 112 EYERHADGRYTSDISSYLEGQAAKEF#IAWLVNGRGRRDFLEEAGTADDIGRRHADGTFT 170 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNI 88 +||+ |||||||++ || |+|#++||+| | + ++ Sbjct 162 RRHADGTFTSDYNQLLDDIATQEF#LKWLINQKVTQRDL 199 |
| Rattus norvegicus | Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| | |||#| ||+ | ||+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDF#VNWLLAQKGKKND--WKHNLTQREA 88 |
| Canis familiaris | Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| | |||#| ||+ | ||+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDF#VNWLLAQKGKKND--WKHNITQREA 88 |
| Ictalurus punctatus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 +|||+|||++||||||++||||||#||||||+||+ + |||+||+| Sbjct 49 RRHSEGTFSNDYSKYLETRRAQDF#VQWLMNSKRSDS-------PARRHADGTYT 95 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEF--ERHAEGTFT 104 +||+ ||+||| | || + |+||#+ || + + + + | + || +|+|| Sbjct 87 RRHADGTYTSDVSSYLQDQAAKDF#ITWLKSGQPKPDAVETRSVDTLRRRHVDGSFT 142 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRH 92 +|| |+|||| +| ||| |+++#+||+||+ + + ||| Sbjct 134 RRHVDGSFTSDVNKVLDSLAAKEY#LQWVMNSPSSAS--GKRH 173 |
| Xenopus tropicalis | Query 46 QMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 |+ | ||||||||||||||||||||||||#+||||||||+ +++|+ ++||||||||| Sbjct 46 QLKEVKRHSQGTFTSDYSKYLDSRRAQDF#IQWLMNTKRS-GGLSRRNADYERHAEGTFT 103 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 +||++|||||| +++|| + |++|#+ ||+| +| |++|+ + |||||||+| Sbjct 95 ERHAEGTFTSDVTQHLDEKAAKEF#IDWLINGGPSKEIISRRNADVERHAEGTYT 148 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 +||++||+|+| ++||+ + |++|#++||+| | | + |||||||| Sbjct 140 ERHAEGTYTNDVTEYLEEKAAKEF#IEWLINGKPKK-------IRYSRHAEGTFT 186 Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFE---RHAEGTFT 104 ||++||||+| + ||+ + |++|#| ||+ | | | ++ | | |||+|+|| Sbjct 179 RHAEGTFTNDMTNYLEEKAAKEF#VGWLIK-GRPKRNFSEAHSVEEMDRRHADGSFT 233 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNI 88 |+||+ |+||+| +| || ||+|#+ |++||+ + ++ Sbjct 224 DRRHADGSFTNDINKVLDIIAAQEF#LDWVINTQVTERDL 262 |
| Capra hircus | Query 86 NNIAKRHDEFERHAEGTFT 104 ||||||||||||||||||| Query 86 NNIAKRHDEFERHAEGTFT 104 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLM 79 +| +||++|||||| | ||+ + |++|#+ ||+ Sbjct 8 DEFERHAEGTFTSDVSSYLEGQAAKEF#IAWLV 39 |
| Bos taurus | Query 47 MNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ++ + ||+ | ||||||+ | |+ +#++ |+ || |+|++ +||++ || Sbjct 75 VSRNVRHADGVFTSDYSRLLGQLSAKKY#LESLIG-KRVSNSISEDQGPIKRHSDAVFT 131 |
| Oncorhynchus tshawytscha | Query 54 SQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 | |||++||||| + | ||||#||||||+||+ +|||+||+| Sbjct 2 SXGTFSNDYSKYQEERMAQDF#VQWLMNSKRS-------GAPSKRHADGTYT 45 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNK 85 |||+ ||+||| | || + |+||#| || + + + Sbjct 37 KRHADGTYTSDVSTYLQDQAAKDF#VSWLKSGRARR 71 |
| Danio rerio | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ||||+|||++||||||++||||||#|||||| ||| ++ +||||||+| Sbjct 48 KRHSEGTFSNDYSKYLETRRAQDF#VQWLMNAKRNGGSV-------KRHAEGTYT 94 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWL 78 |||++||+||| | || + || |#| || Sbjct 86 KRHAEGTYTSDVSSYLQDQAAQSF#VAWL 113 |
| Macaca mulatta | Query 47 MNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ++ + ||+ | ||||+|| | |+ +#++ || || +||++ +||++ || Sbjct 75 VSRNARHADGVFTSDFSKLLGQLSAKKY#LESLMG-KRVSSNISEDPVPVKRHSDAVFT 131 |
| Pan troglodytes | Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| |||#| ||+ | ||+ +|+ +| | Sbjct 51 RYAEGTFISDYSIAMDKIHQQDF#VNWLLAQKGKKND--WKHNITQREA 96 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRN 84 |+ ||||+| + |||++ |+||#| || + ++ Sbjct 1 HADGTFTNDMTSYLDAKAARDF#VSWLARSDKS 32 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ||||+|||++||||||++||||||#|||| |+||| | |||+||+| Sbjct 50 KRHSEGTFSNDYSKYLETRRAQDF#VQWLKNSKRN--------GLFRRHADGTYT 95 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWL 78 +||+ ||+||| | || + |+||#| || Sbjct 87 RRHADGTYTSDVSSYLQDQAAKDF#VSWL 114 |
| Mus musculus | Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| | |||#| ||+ + |++ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDF#VNWLLAQRGKKSD--WKHNITQREA 88 |
| Sebastes caurinus | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 |+++ ||||+|||++|||+||+ |+||||#|+|||| ||+ || +|||+|||| Sbjct 42 DELSNMKRHSEGTFSNDYSRYLEERKAQDF#VRWLMNNKRS------GADE-KRHADGTFT 94 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK-RNKNNIAKRHDEF-ERHAEGTFT 104 ++|||+ |||||| | || + +||#| | + + | ++ | + | || +|+|| Sbjct 84 DEKRHADGTFTSDVSSYLKDQAIKDF#VNRLKSGQVRRESETEWRGEAFSRRHVDGSFT 141 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDE 94 +|| |+|||| +| ||| |+++#+ |+| +| + + ++ |+ Sbjct 133 RRHVDGSFTSDVNKVLDSMAAKEY#LLWVMTSKPSGESKKRQEDQ 176 |
| Heloderma suspectum | Query 46 QMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKR------------------NKNN 87 |+|| |||||||||||||||||+||||||#||||||||| + | Sbjct 46 QLNEVKRHSQGTFTSDYSKYLDTRRAQDF#VQWLMNTKRSGQQGVEEREKENLLDQLSSNG 105 Query 88 IAKRHDEFERHAEGTFT 104 +|+ | |+||||+| +| Sbjct 106 LARHHAEYERHADGRYT 122 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMN 80 | +||+ | +||| | ||+ + |++|#+ ||+| Sbjct 112 EYERHADGRYTSDISSYLEGQAAKEF#IAWLVN 143 |
| Squalus acanthias | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWL--MNTKRNKNNIAKRHDEFERHAEGTF 103 ++ |||++||+||| | +|+ |#| | | ++| +|+||| | || ||++ Sbjct 20 DKTKRHAEGTYTSDVDSLSDYFKAKRF#VDSLTSYNKRQNGRSISKRHVESTRHTEGSY 77 Query 67 DSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 |+|||+||#|||||+|||| |+ +||||||+| Sbjct 1 DNRRAKDF#VQWLMSTKRN-------GDKTKRHAEGTYT 31 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFE 96 |+||+ |||||+ + ||+ |++|#+ |++|+| | | | | Sbjct 122 DRRHADGTFTSEINIVLDTIAAKEF#LNWILNSK----TIQSRDSEIE 164 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKN---------NIAKRHDEFERHA 99 | ||++|++ | | ||+++ |+||#+ ||+ + + | + ||| Sbjct 68 ESTRHTEGSYR-DISSYLEAKAAKDF#INWLIKGRGRREFPEESKEIVNEVIPEEMDRRHA 126 Query 100 EGTFT 104 +|||| Sbjct 127 DGTFT 131 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ||||+|||++||||||++||||||#|+||||+| | + +||||||+| Sbjct 48 KRHSEGTFSNDYSKYLETRRAQDF#VEWLMNSKSNGGSA-------KRHAEGTYT 94 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWL 78 |||++||+||| | +| + ||+|#| || Sbjct 86 KRHAEGTYTSDISSFLRDQAAQNF#VAWL 113 |
| Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 || |+|||++ |||| + | |+||#+ ||||+||+ +|||+||+| Sbjct 50 KRQSEGTFSNYYSKYQEERMARDF#LHWLMNSKRS-------GAPSKRHADGTYT 96 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMN--TKRNKNNIAKRHDEFERHAEGTFT 104 |||+ ||+||| | || + |+||#| || + +| + || +|+|| Sbjct 88 KRHADGTYTSDVSTYLQDQAAKDF#VSWLKSGPARRESAEESMNGPMSRRHVDGSFT 143 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHD 93 +|| |+|||| +| ||| |+++#+ |+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSLAAKEY#LLWVMTSKTSGKSNKRQED 177 |
| Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ||||+|||++||||| + | ||||#||||||+||+ +|||+||+| Sbjct 50 KRHSEGTFSNDYSKYQEERMAQDF#VQWLMNSKRS-------GAPSKRHADGTYT 96 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMN--TKRNKNNIAKRHDEFERHAEGTFT 104 |||+ ||+||| | || + |+||#| || + +| ++ || +|+|| Sbjct 88 KRHADGTYTSDVSTYLQDQAAKDF#VSWLKSGRARRESAEESRNGPMSRRHVDGSFT 143 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHD 93 +|| |+|||| +| ||| |+++#+ |+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSLAAKEY#LLWVMTSKTSGKSNKRQED 177 |
| Heloderma horridum | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMN 80 |||| |||||| || ++ + |#++|| | Sbjct 46 KRHSDGTFTSDLSKQMEEEAVRLF#IEWLKN 75 |
| Tetraodon nigroviridis | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ||||+|||++||||||++||||||#|||| |+||| + | |||+||+| Sbjct 787 KRHSEGTFSNDYSKYLETRRAQDF#VQWLKNSKRNGS-------LFRRHADGTYT 833 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWL 78 +||+ ||+||| | || + |++|#| || Sbjct 825 RRHADGTYTSDVSAYLQDQAAKEF#VSWL 852 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 |+ |+|||| +| ||+ |++|#+ ||++|| Sbjct 1 HADGSFTSDINKVLDTIAAKEF#LNWLISTK 30 |
| Protopterus dolloi | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 | |||||||| |||+| |+++ | | #|| ||+||| ++|||+| |||||||+| Sbjct 28 EAKRHSQGTFMSDYAKLLEAKHALDL#VQRLMSTKRT-GAVSKRHNEIERHAEGTYT 82 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFE-RHAEGTFT 104 || +||++||+||| | ||+ + +|#++||| + ++ +| + |||+|| | Sbjct 71 NEIERHAEGTYTSDISSYLEGQAVNEF#IRWLMKGRGRRDFSETDIEEMDRRHADGTVT 128 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 |+||+ || ||| + |+| ++|#+ ||+| | Sbjct 119 DRRHADGTVTSDINSVLESIATREF#LNWLINAK 151 |
| Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | Query 47 MNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ++ + ||+ | ||||||+ | |+ +#++ |+ || ++|++ +||++ || Sbjct 75 VSRNARHADGVFTSDYSRLLGQISAKKY#LESLIG-KRISSSISEDPVPIKRHSDAVFT 131 |
| Ambloplites rupestris | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 |||||||+||+ ||+ |+||||#++|||| || + |++ |||+|||| Sbjct 1 HSQGTFTNDYTNYLEDRQAQDF#IRWLMNNKR--SGAAEK-----RHADGTFT 45 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK-RNKNNIAKRHDEF-ERHAEGTFT 104 +|||+ |||| | | + +||#| | + + | + +| + | || ||+|| Sbjct 36 EKRHADGTFTDDASSDFYDQAIKDF#VAKLKSGQDRREPETDRRREAFSRRHVEGSFT 92 |
| Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | Query 52 RHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 |||||+|||||||+|| ++|+||#| ||+|||| + + +||++|+|| Sbjct 43 RHSQGSFTSDYSKHLDVKQAKDF#VTWLLNTKRRGVDAQAGANLEKRHSDGSFT 95 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKR 83 | +|||| |+||+| + || |++|#++|| | Sbjct 84 NLEKRHSDGSFTNDMNVMLDRMSAKNF#LEWLKQQGR 119 |
| Arvicanthis ansorgei | Query 47 MNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ++ + ||+ | ||||||+ | |+ +#++ |+ || ++| + +||++ || Sbjct 58 VSRNARHADGVFTSDYSRLLGQISAKKY#LESLIG-KRISSSILEDPVPIKRHSDAVFT 114 |
| synthetic construct | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ||||||||||||||||||||||||||||||#||||||||||+||||||||||||||||||| Sbjct 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNRNNIAKRHDEFERHAEGTFT 104 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKN---NIAKRHDEFERHAEGTFT 104 +| +||++|||||| | ||+ + |++|#+ ||+ + ++ +| + |||+|+|+ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQAAKEF#IAWLVKGRGRRDFPEEVAIVEELGRRHADGSFS 152 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 +||+ |+|+ + + ||+ |+||#+ ||+ || Sbjct 144 RRHADGSFSDEMNTILDNLAARDF#INWLIQTK 175 |
| Phodopus sungorus | Query 56 GTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKN---NIAKRHDEFERHAEGTFT 104 |||||| | ||+ + |++|#+ ||+ + ++ + + |||+|+|+ Sbjct 1 GTFTSDVSSYLEGQAAKEF#IAWLVKGRGRRDFPEEVTIVEELGRRHADGSFS 52 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 +||+ |+|+ + + ||| +||#+ ||+ || Sbjct 44 RRHADGSFSDEMNTILDSLATRDF#INWLIQTK 75 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | Query 46 QMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 +++ ||||+|||++||||||+ |+||+|#|+|||| || + +|++ |||+|||| Sbjct 46 ELSNMKRHSEGTFSNDYSKYLEDRKAQEF#VRWLMNNKR--SGVAEK-----RHADGTFT 97 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDF#VQWL 78 +|||+ |||||| | || + +||#| | Sbjct 88 EKRHADGTFTSDVSSYLKDQAIKDF#VDRL 116 |
| Sus scrofa | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ||| ||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 45 DQMTEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKN---NIAKRHDEFERHAEGTFT 104 +| +||++|||||| | ||+ + |++|#+ ||+ + ++ + + |||+|+|+ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQAAKEF#IAWLVKGRGRRDFPEEVTIVEELRRRHADGSFS 152 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 +||+ |+|+ + + ||+ +||#+ ||++|| Sbjct 144 RRHADGSFSDEMNTVLDNLATRDF#INWLLHTK 175 |
| Neoceratodus forsteri | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 | |||||||| |||+| |++| | ||#|| ||||||| ++||| | |||||||+| Sbjct 20 EAKRHSQGTFMSDYAKLLEARHALDF#VQRLMNTKRN-GGVSKRHSEIERHAEGTYT 74 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEF-ERHAEGTFT 104 +| +||++||+||| | ||+ + | +|#|+||+ + ++ +| |||+|| | Sbjct 63 SEIERHAEGTYTSDISSYLEGQAANEF#VRWLLKGRGRRDFSDTDAEEMGRRHADGTIT 120 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 +||+ || |+| + |+| ++|#+ ||+|+| Sbjct 112 RRHADGTITNDMNNVLESIATREF#LNWLINSK 143 |
| Canis lupus familiaris | Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 45 DQMNEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKN---NIAKRHDEFERHAEGTFT 104 +| +||++|||||| | ||+ + |++|#+ ||+ + ++ +| + |||+|+|+ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQAAKEF#IAWLVKGRGRRDFPEEVAIVEEFRRRHADGSFS 152 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTK 82 +||+ |+|+ + + ||+ +||#+ ||+ || Sbjct 144 RRHADGSFSDEMNTVLDTLATRDF#INWLLQTK 175 |
| Heloderma suspectum, venom, Peptide, 39 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMN 80 | +|||||| || ++ + |#++|| | Sbjct 1 HGEGTFTSDLSKQMEEEAVRLF#IEWLKN 28 |
| Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 |||||||+|||||||+||||||#|||||+|||+ | ||+||+| Sbjct 1 HSQGTFTNDYSKYLDTRRAQDF#VQWLMSTKRS-GGITXXXXXXXXHADGTYT 51 Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWL 78 |+ ||+||| | || + |+ |#| || Sbjct 45 HADGTYTSDVSSYLQDQAAKKF#VTWL 70 |
| Hydrolagus colliei | Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 |||+ | |+||||||||+|| +||#||||++|||| | |+ +|||+| +| Sbjct 30 KRHTDGIFSSDYSKYLDNRRTKDF#VQWLLSTKRNGANT----DKTKRHADGIYT 79 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNN--IAKRHDEFERHAE 100 ++ |||+ | +||| + | +++ |#|+ | | + +|+ ++||| | | | Sbjct 68 DKTKRHADGIYTSDVASLTDYLKSKRF#VESLSNYNKRQNDRRMSKRHVEGARRTE 122 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFE 96 | | ++ + +|+| ||+++ |+||#+ ||+ + + + |+ | | Sbjct 116 EGARRTEEDYPNDFSSYLEAKAAKDF#IDWLIK-GQGRRDFAEESREIE 162 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDF#VQWLMN 80 |+ || ||| | +|+ + ++|#+ ||++ Sbjct 1 HADGTLTSDISSFLEKQATKEF#IAWLVS 28 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | Query 56 GTFTSDYSKYLDSRRAQDF#VQWL 78 ||+||| | || + |++|#| || Sbjct 3 GTYTSDVSSYLQDQAAKEF#VSWL 25 |
| Agkistrodon piscivorus | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNIAKRH---DEFERHAEGTFT 104 | +||+ ||+||| | ||+ + |++|#+ ||+| + ++ | | |||+|||| Sbjct 32 EYERHADGTYTSDISSYLEGQAAKEF#IAWLVNGRGRRDFSEGAHTGEDLGRRHADGTFT 90 Query 51 KRHSQGTFTSDYSKYLDSRRAQDF#VQWLMNTKRNKNNI 88 +||+ |||||||+| || |+|#++||+| | + ++ Sbjct 82 RRHADGTFTSDYNKLLDDIATQEF#LKWLINQKVTQRDL 119 Query 66 LDSRRAQDF#VQWLMNTKRNKNNIAKRHDEFERHAEGTFT 104 |+ | ++|#+ | + | +|+ | |+||||+||+| Sbjct 9 LEDREKENF#LDQLAS-----NGLARPHAEYERHADGTYT 42 |
[Site 2] SDYSKYLDSR69-RAQDFVQWLM
Arg69  Arg
Arg
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser60 | Asp61 | Tyr62 | Ser63 | Lys64 | Tyr65 | Leu66 | Asp67 | Ser68 | Arg69 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Arg70 | Ala71 | Gln72 | Asp73 | Phe74 | Val75 | Gln76 | Trp77 | Leu78 | Met79 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| PLSDPDQMNEDKRHSQGTFTSDYSKYLDSRRAQDFVQWLMNTKRNKNNIAKRHDEFERHA |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 129.00 | 10 | glucagon, isoform CRA_b |
| 2 | Pan troglodytes | 102.00 | 2 | PREDICTED: fibroblast activation protein, alpha su |
| 3 | N/A | 80.90 | 44 | 0 |
| 4 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | 64.70 | 1 | AAB31092.1 |
| 5 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | 61.60 | 1 | glucagon=proglucagon-derived 3.5 kda peptide |
| 6 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | 60.80 | 1 | glucagon II |
| 7 | Contains: Glucagon; Glucagon-like peptide | 60.10 | 4 | GLUC_AMICA Glucagon precursor |
| 8 | Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | 60.10 | 1 | glucagon |
| 9 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | 58.50 | 1 | glucagon G-29 |
| 10 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | 57.80 | 1 | GLUC1_ORENI Glucagon-1 (Glucagon I) gb |
| 11 | Ambloplites rupestris | 57.40 | 1 | proglucagon |
| 12 | Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | 56.20 | 1 | GLUC_LAMFL Glucagon gb |
| 13 | deshis1, Desphe6, Glu9 | 53.50 | 1 | A Chain A, Nmr Solution Structure Of The Glucagon |
| 14 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | 53.50 | 1 | glucagon |
| 15 | Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | 46.60 | 1 | GLUC1_PETMA Glucagon-1 precursor |
| 16 | Monodelphis domestica | 45.10 | 3 | PREDICTED: similar to Gastric inhibitory polypepti |
| 17 | Rattus norvegicus | 43.90 | 5 | Glucose-dependent insulinotropic peptide |
| 18 | Canis familiaris | 43.90 | 2 | PREDICTED: similar to Gastric inhibitory polypepti |
| 19 | Agkistrodon piscivorus | 43.90 | 1 | proglucagon |
| 20 | Xenopus laevis | 43.50 | 5 | proglucagon I |
| 21 | Gallus gallus | 43.10 | 4 | glucagon isoform 1 |
| 22 | Meleagris gallopavo | 43.10 | 2 | preproglucagon B |
| 23 | Xenopus tropicalis | 42.70 | 2 | glucagon |
| 24 | Hoplobatrachus rugulosus | 42.70 | 1 | AF324209_1 proglucagon |
| 25 | Oncorhynchus tshawytscha | 42.40 | 1 | S78474_1 proglucagon |
| 26 | Danio rerio | 42.00 | 5 | hypothetical protein LOC494052 |
| 27 | Macaca mulatta | 42.00 | 3 | PREDICTED: gastric inhibitory polypeptide |
| 28 | Bos taurus | 41.60 | 5 | PREDICTED: similar to Gastric inhibitory polypepti |
| 29 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | 41.20 | 1 | glucagon-like peptide, GLP |
| 30 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | 41.20 | 1 | GLUC2_LOPAM Glucagon-2 precursor (Glucagon II) |
| 31 | Mus musculus | 40.80 | 6 | gastric inhibitory polypeptide |
| 32 | Ictalurus punctatus | 40.80 | 3 | proglucagon I |
| 33 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | 40.80 | 1 | GLUC_HELSU Glucagon precursor |
| 34 | Heloderma suspectum | 40.40 | 2 | proglucagon |
| 35 | Protopterus dolloi | 40.40 | 1 | proglucagon |
| 36 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | 40.00 | 2 | GLUC_CARAU Glucagon precursor |
| 37 | Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC2_ONCMY Glucagon-2 precursor (Glucagon II) |
| 38 | Heloderma horridum | 40.00 | 1 | EXE3_HELHO Exendin-3 precursor emb |
| 39 | Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC1_ONCMY Glucagon-1 precursor (Glucagon I) |
| 40 | Neoceratodus forsteri | 39.70 | 1 | proglucagon |
| 41 | Sebastes caurinus | 39.30 | 4 | proglucagon II |
| 42 | Tetraodon nigroviridis | 39.30 | 2 | unnamed protein product |
| 43 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | 38.90 | 1 | glucagon-like peptide-2, GLP-2=proglucagon-derived |
| 44 | Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | 38.10 | 1 | GLUC2_PETMA Glucagon-2 precursor |
| 45 | synthetic construct | 37.00 | 1 | Homo sapiens glucagon |
| 46 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | 36.60 | 1 | GLUC1_LOPAM Glucagon-1 precursor (Glucagon I) |
| 47 | Sus scrofa | 36.20 | 1 | glucagon |
| 48 | Canis lupus familiaris | 35.80 | 1 | glucagon |
| 49 | Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | 35.40 | 1 | GLUC_LEPSP Glucagon precursor |
| 50 | Phodopus sungorus | 35.40 | 1 | glucagon precursor |
| 51 | Heloderma suspectum, venom, Peptide, 39 aa | 35.40 | 1 | A Chain A, Solution Structure Of Exendin-4 In 30-V |
| 52 | Bufo marinus | 35.40 | 1 | proglucagon |
| 53 | Hydrolagus colliei | 34.30 | 1 | proglucagon |
| 54 | Squalus acanthias | 34.30 | 1 | proglucagon |
| 55 | Capra hircus | 34.30 | 1 | glucagon |
| 56 | Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | 33.90 | 1 | VIP_MOUSE VIP peptides precursor |
| 57 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | 33.50 | 1 | GLUC2_ORENI Glucagon-2 (Glucagon II) gb |
| 58 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | 33.50 | 1 | glucagon-like peptide-1, GLP-1=proglucagon-derived |
| 59 | Arvicanthis ansorgei | 33.10 | 1 | vasoactive intestinal polypeptide |
| 60 | Candida glabrata | 33.10 | 1 | unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 ||||||||||||||||||||||||||||||#|||||||||||||||+|||||||||||||| Sbjct 123 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 182 |
| Pan troglodytes | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDE 94 ||||||||||||||||||||||||||||||#||||||||||||||| +++| || | Sbjct 962 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN-SSVAYRHAE 1015 |
| N/A | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIA 89 |||||||||||||||||#|||||||||||||||||||| Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIA 89 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN 84 ||+||||||||||+|+|#||+|||||||+|||| Sbjct 1 HSEGTFTSDYSKYMDNR#RAKDFVQWLMSTKRN 32 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 |||||||||||||||+|#|||||+||||+| Sbjct 1 HSQGTFTSDYSKYLDNR#RAQDFIQWLMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN 84 ||+|||++||||||++|#||||||||| |+||| Sbjct 1 HSEGTFSNDYSKYLETR#RAQDFVQWLKNSKRN 32 |
| Contains: Glucagon; Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 |||||||+|||||+|+|#||||||||||+| Sbjct 1 HSQGTFTNDYSKYMDTR#RAQDFVQWLMST 29 |
| Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 |||||||+|||||+|+|#||||||||||+| Sbjct 1 HSQGTFTNDYSKYMDTR#RAQDFVQWLMST 29 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 ||+||||||||||+|+|#||+|||||||+| Sbjct 1 HSEGTFTSDYSKYMDNR#RAKDFVQWLMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN 84 ||+|||++||||||+ |#+|||||+|||| ||+ Sbjct 1 HSEGTFSNDYSKYLEDR#KAQDFVRWLMNNKRS 32 |
| Ambloplites rupestris | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRH 92 |||||||+||+ ||+ |#+||||++|||| ||+ ||| Sbjct 1 HSQGTFTNDYTNYLEDR#QAQDFIRWLMNNKRS-GAAEKRH 39 |
| Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 ||||+|||||||||||+#+|+||| ||||| Sbjct 1 HSQGSFTSDYSKYLDSK#QAKDFVIWLMNT 29 |
| deshis1, Desphe6, Glu9 | Query 54 SQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 |||| ||+||||||||#|||||||||||| Sbjct 1 SQGT-TSEYSKYLDSR#RAQDFVQWLMNT 27 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMN 80 ||+|||||||||||+++#+|+|||+|||| Sbjct 1 HSEGTFTSDYSKYLENK#QAKDFVRWLMN 28 |
| Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRH 92 ||||+|||||||||||+++#+|+|||+|||| || + + +|| Sbjct 41 KRHSEGTFTSDYSKYLENK#QAKDFVRWLMNAKRGGSELQRRH 82 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLM---NTKRNKNNIAKRHDEFERHA 99 +||+ ||||+| + |||++# |+||| || ++|+ + + | +||| Sbjct 80 RRHADGTFTNDMTSYLDAK#AARDFVSWLARSDKSRRDGGDHLAENSEDKRHA 131 |
| Monodelphis domestica | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 +|+++||| |||| +| # |||| ||++ | || + ||+ || | Sbjct 51 QRYAEGTFISDYSITMDKI#MQQDFVNWLLSQKGKKN--SWRHNITERKA 97 |
| Rattus norvegicus | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| #| |||| ||+ | ||+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKI#RQQDFVNWLLAQKGKKND--WKHNLTQREA 88 |
| Canis familiaris | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| #| |||| ||+ | ||+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKI#RQQDFVNWLLAQKGKKND--WKHNITQREA 88 |
| Agkistrodon piscivorus | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNI 88 +||+ |||||||+| || # |+|++||+| | + ++ Sbjct 82 RRHADGTFTSDYNKLLDDI#ATQEFLKWLINQKVTQRDL 119 Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRH---DEFERHA 99 | +||+ ||+||| | ||+ +# |++|+ ||+| + ++ | | ||| Sbjct 32 EYERHADGTYTSDISSYLEGQ#AAKEFIAWLVNGRGRRDFSEGAHTGEDLGRRHA 85 |
| Xenopus laevis | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 + | +|+ | |||||||||||||||||||#||||||||||||||+ +++|+ ++|||| Sbjct 41 VDDSEQLKEVKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRS-GELSRRNADYERHA 98 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 +||++|||||| ++ || +# |++|+ ||+| +| |++|+ |||||| Sbjct 95 ERHAEGTFTSDVTQQLDEK#AAKEFIDWLINGGPSKEIISRRNAEFERHA 143 Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLM--NTKRNKNNIAKRHDEFERHA 99 ||++||||+| + ||+ +# |++|| ||+ ||| + + + ||| Sbjct 179 RHAEGTFTNDMTNYLEEK#AAKEFVGWLIKGRPKRNFSEVHSVEEMDRRHA 228 Query 50 DKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNI 88 |+||+ |+||+| +| || # ||+|+ |++||+ + ++ Sbjct 224 DRRHADGSFTNDINKVLDII#AAQEFLDWVINTQETERDL 262 Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDE 94 | +||++||+|+| ++||+ +# |++|++||+ | | + || | Sbjct 138 EFERHAEGTYTNDVTEYLEEK#AAKEFIEWLIKGKPKKIRYS-RHAE 182 |
| Gallus gallus | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN--------------- 84 || + |+|| |||||||||||||||||||#||||||||||+|||| Sbjct 42 PLDESRQLNEVKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMSTKRNGQQGQEDKENDKFPD 101 Query 85 ---KNNIAKRHDEFERHA 99 | |+||| |||||| Sbjct 102 QLSSNAISKRHSEFERHA 119 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNI 88 +||+ |||||| +| || # |++|++||+||| + ++ Sbjct 164 RRHADGTFTSDINKILDDM#AAKEFLKWLINTKVTQRDL 201 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKN 86 +| +||++||+||| + ||+ +# |++|+ ||+| + ++ Sbjct 113 SEFERHAEGTYTSDITSYLEGQ#AAKEFIAWLVNGRGRRD 151 |
| Meleagris gallopavo | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN--------------- 84 || + |+|| |||||||||||||||||||#||||||||||+|||| Sbjct 42 PLDESRQLNEVKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMSTKRNGQQGQEDKENDEFPD 101 Query 85 ---KNNIAKRHDEFERHA 99 | |+||| |||||| Sbjct 102 QLSSNAISKRHSEFERHA 119 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNI 88 +||+ |||||| +| || # |++|++||+||| + ++ Sbjct 164 RRHADGTFTSDINKILDDM#AAKEFLKWLINTKVTQRDL 201 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKN 86 +| +||++||+||| + ||+ +# |++|+ ||+| + ++ Sbjct 113 SEFERHAEGTYTSDITSYLEGQ#AAKEFIAWLVNGRGRRD 151 |
| Xenopus tropicalis | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 + | |+ | |||||||||||||||||||#|||||+||||||||+ +++|+ ++|||| Sbjct 41 VDDSQQLKEVKRHSQGTFTSDYSKYLDSR#RAQDFIQWLMNTKRS-GGLSRRNADYERHA 98 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 +||++|||||| +++|| +# |++|+ ||+| +| |++|+ + |||| Sbjct 95 ERHAEGTFTSDVTQHLDEK#AAKEFIDWLINGGPSKEIISRRNADVERHA 143 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDE 94 +||++||+|+| ++||+ +# |++|++||+| | | + || | Sbjct 140 ERHAEGTYTNDVTEYLEEK#AAKEFIEWLINGKPKKIRYS-RHAE 182 Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFE---RHA 99 ||++||||+| + ||+ +# |++|| ||+ | | | ++ | | ||| Sbjct 179 RHAEGTFTNDMTNYLEEK#AAKEFVGWLIK-GRPKRNFSEAHSVEEMDRRHA 228 Query 50 DKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNI 88 |+||+ |+||+| +| || # ||+|+ |++||+ + ++ Sbjct 224 DRRHADGSFTNDINKVLDII#AAQEFLDWVINTQVTERDL 262 |
| Hoplobatrachus rugulosus | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 | | | || |||||||||||||||||||#|||||||||||+||+ |+||+ +||||| Sbjct 41 LDDSDHHNEVKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNSKRS-GGISKRNVQFERHA 98 Query 50 DKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 |+||+ |+||||++| || +# ||+|+ |++|| Sbjct 178 DRRHADGSFTSDFNKALDIK#AAQEFLDWIINT 209 Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKN 86 ||++|||||| + ||+ +# |++|| ||+ + +| Sbjct 134 RHAEGTFTSDMTSYLEEK#AAKEFVDWLIKGRPKRN 168 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDE 94 +||++||+|+| +++|+ +# |++|+ ||+ | | ++ || | Sbjct 95 ERHAEGTYTNDVTQFLEEK#AAKEFIDWLIKGKPKKQRLS-RHAE 137 |
| Oncorhynchus tshawytscha | Query 54 SQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRH 92 | |||++||||| + |# ||||||||||+||+ +||| Sbjct 2 SXGTFSNDYSKYQEER#MAQDFVQWLMNSKRS-GAPSKRH 39 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNK 85 |||+ ||+||| | || +# |+||| || + + + Sbjct 37 KRHADGTYTSDVSTYLQDQ#AAKDFVSWLKSGRARR 71 |
| Danio rerio | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDE 94 ||||+|||++||||||++|#||||||||||| ||| ++ ||| | Sbjct 48 KRHSEGTFSNDYSKYLETR#RAQDFVQWLMNAKRNGGSV-KRHAE 90 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWL 78 |||++||+||| | || +# || || || Sbjct 86 KRHAEGTYTSDVSSYLQDQ#AAQSFVAWL 113 |
| Macaca mulatta | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| # |||| ||+ | ||+ +|+ +| | Sbjct 51 RYAEGTFISDYSIAMDKI#HQQDFVNWLLAQKGKKND--WKHNITQREA 96 |
| Bos taurus | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNN 87 |+++||| |||| +| #| |||| ||+ | |++ Sbjct 81 RYAEGTFISDYSIAMDKI#RQQDFVNWLLAQKGKKSD 116 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN 84 |+ ||||+| + |||++# |+||| || + ++ Sbjct 1 HADGTFTNDMTSYLDAK#AARDFVSWLARSDKS 32 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRH 92 |++| + ||||+|||++||||||++|#||||||||| |+|| | + +|| Sbjct 40 LTEPIEPLNMKRHSEGTFSNDYSKYLETR#RAQDFVQWLKNSKR--NGLFRRH 89 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWL 78 +||+ ||+||| | || +# |+||| || Sbjct 87 RRHADGTYTSDVSSYLQDQ#AAKDFVSWL 114 |
| Mus musculus | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| #| |||| ||+ + |++ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKI#RQQDFVNWLLAQRGKKSD--WKHNITQREA 88 |
| Ictalurus punctatus | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRH 92 +|||+|||++||||||++|#|||||||||||+||+ ++ |+|| Sbjct 49 RRHSEGTFSNDYSKYLETR#RAQDFVQWLMNSKRS-DSPARRH 89 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRH 92 +|| |+|||| +| ||| # |++++||+||+ + + ||| Sbjct 134 RRHVDGSFTSDVNKVLDSL#AAKEYLQWVMNSPSSAS--GKRH 173 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKR 91 +||+ ||+||| | || +# |+||+ || + + + + | Sbjct 87 RRHADGTYTSDVSSYLQDQ#AAKDFITWLKSGQPKPDAVETR 127 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKR---------------- 83 || | |+|| |||||||||||||||||+|#|||||||||||||| Sbjct 40 PLDDSRQLNEVKRHSQGTFTSDYSKYLDTR#RAQDFVQWLMNTKRSGQQGVEEREKENLLD 99 Query 84 --NKNNIAKRHDEFERHA 99 + | +|+ | |+|||| Sbjct 100 QLSSNGLARHHAEYERHA 117 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNI 88 +||+ |||||||++ || # |+|++||+| | + ++ Sbjct 162 RRHADGTFTSDYNQLLDDI#ATQEFLKWLINQKVTQRDL 199 Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKN 86 | +||+ | +||| | ||+ +# |++|+ ||+| + ++ Sbjct 112 EYERHADGRYTSDISSYLEGQ#AAKEFIAWLVNGRGRRD 149 |
| Heloderma suspectum | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKR---------------- 83 || | |+|| |||||||||||||||||+|#|||||||||||||| Sbjct 40 PLDDSRQLNEVKRHSQGTFTSDYSKYLDTR#RAQDFVQWLMNTKRSGQQGVEEREKENLLD 99 Query 84 --NKNNIAKRHDEFERHA 99 + | +|+ | |+|||| Sbjct 100 QLSSNGLARHHAEYERHA 117 Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMN 80 | +||+ | +||| | ||+ +# |++|+ ||+| Sbjct 112 EYERHADGRYTSDISSYLEGQ#AAKEFIAWLVN 143 |
| Protopterus dolloi | Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 | |||||||| |||+| |+++# | | || ||+||| ++|||+| |||| Sbjct 28 EAKRHSQGTFMSDYAKLLEAK#HALDLVQRLMSTKRT-GAVSKRHNEIERHA 77 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFER 97 || +||++||+||| | ||+ +# +|++||| + ++ +| +| Sbjct 71 NEIERHAEGTYTSDISSYLEGQ#AVNEFIRWLMKGRGRRDFSETDIEEMDR 120 Query 42 SDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 |+ | |+||+ || ||| + |+| # ++|+ ||+| | Sbjct 111 SETDIEEMDRRHADGTVTSDINSVLESI#ATREFLNWLINAK 151 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDE 94 ||||+|||++||||||++|#||||||+||||+| | + |||| | Sbjct 48 KRHSEGTFSNDYSKYLETR#RAQDFVEWLMNSKSNGGS-AKRHAE 90 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWL 78 |||++||+||| | +| +# ||+|| || Sbjct 86 KRHAEGTYTSDISSFLRDQ#AAQNFVAWL 113 |
| Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRH 92 || |+|||++ |||| + |# |+||+ ||||+||+ +||| Sbjct 50 KRQSEGTFSNYYSKYQEER#MARDFLHWLMNSKRS-GAPSKRH 90 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWL 78 |||+ ||+||| | || +# |+||| || Sbjct 88 KRHADGTYTSDVSTYLQDQ#AAKDFVSWL 115 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHD 93 +|| |+|||| +| ||| # |++++ |+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSL#AAKEYLLWVMTSKTSGKSNKRQED 177 |
| Heloderma horridum | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMN 80 |||| |||||| || ++ # + |++|| | Sbjct 46 KRHSDGTFTSDLSKQMEEE#AVRLFIEWLKN 75 |
| Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRH 92 | | ++ ||||+|||++||||| + |# ||||||||||+||+ +||| Sbjct 40 LEDLMGVSNVKRHSEGTFSNDYSKYQEER#MAQDFVQWLMNSKRS-GAPSKRH 90 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNN 87 |||+ ||+||| | || +# |+||| || + + + + Sbjct 88 KRHADGTYTSDVSTYLQDQ#AAKDFVSWLKSGRARRES 124 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHD 93 +|| |+|||| +| ||| # |++++ |+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSL#AAKEYLLWVMTSKTSGKSNKRQED 177 |
| Neoceratodus forsteri | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 |+ + | |||||||| |||+| |++|# | |||| ||||||| ++||| | |||| Sbjct 11 PVDASREHTEAKRHSQGTFMSDYAKLLEAR#HALDFVQRLMNTKRN-GGVSKRHSEIERHA 69 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKN 86 +| +||++||+||| | ||+ +# | +||+||+ + ++ Sbjct 63 SEIERHAEGTYTSDISSYLEGQ#AANEFVRWLLKGRGRRD 101 Query 42 SDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 || | +||+ || |+| + |+| # ++|+ ||+|+| Sbjct 103 SDTDAEEMGRRHADGTITNDMNNVLESI#ATREFLNWLINSK 143 |
| Sebastes caurinus | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRH 92 |++| ++ ||||+|||++||||||++|#||||||||| |+||| ++ +|| Sbjct 40 LAEPIELPNMKRHSEGTFSNDYSKYLETR#RAQDFVQWLKNSKRN-GSLFRRH 90 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWL 78 +||+ ||+||| | || +# |++|| || Sbjct 88 RRHADGTYTSDVSSYLQDQ#AAKEFVYWL 115 |
| Tetraodon nigroviridis | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRH 92 |++| ++ ||||+|||++||||||++|#||||||||| |+||| ++ +|| Sbjct 777 LTEPSDLSNMKRHSEGTFSNDYSKYLETR#RAQDFVQWLKNSKRN-GSLFRRH 827 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWL 78 +||+ ||+||| | || +# |++|| || Sbjct 825 RRHADGTYTSDVSAYLQDQ#AAKEFVSWL 852 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 |+ |+|||| +| ||+ # |++|+ ||++|| Sbjct 1 HADGSFTSDINKVLDTI#AAKEFLNWLISTK 30 |
| Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN------KNNIAKRHDE 94 |||||+|||||||+|| +#+|+||| ||+|||| |+ ||| + Sbjct 43 RHSQGSFTSDYSKHLDVK#QAKDFVTWLLNTKRRGVDAQAGANLEKRHSD 91 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKR 83 | +|||| |+||+| + || # |++|++|| | Sbjct 84 NLEKRHSDGSFTNDMNVMLDRM#SAKNFLEWLKQQGR 119 |
| synthetic construct | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 ||||||||||||||||||||||||||||||#|||||||||||||||+|||||||||||||| Sbjct 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNRNNIAKRHDEFERHA 99 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 +||+ |+|+ + + ||+ # |+||+ ||+ || Sbjct 144 RRHADGSFSDEMNTILDNL#AARDFINWLIQTK 175 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | Query 43 DPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRH 92 +| +++ ||||+|||++||||||+ |#+||+||+|||| ||+ ||| Sbjct 43 EPRELSNMKRHSEGTFSNDYSKYLEDR#KAQEFVRWLMNNKRS-GVAEKRH 91 Query 50 DKRHSQGTFTSDYSKYLDSR#RAQDFVQWL 78 +|||+ |||||| | || +# +||| | Sbjct 88 EKRHADGTFTSDVSSYLKDQ#AIKDFVDRL 116 |
| Sus scrofa | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 || ||||| |||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 40 PLDDPDQMTEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 +||+ |+|+ + + ||+ # +||+ ||++|| Sbjct 144 RRHADGSFSDEMNTVLDNL#ATRDFINWLLHTK 175 |
| Canis lupus familiaris | Query 40 PLSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 ||+| |||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 40 PLNDLDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKN 86 +| +||++|||||| | ||+ +# |++|+ ||+ + ++ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQ#AAKEFIAWLVKGRGRRD 131 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 +||+ |+|+ + + ||+ # +||+ ||+ || Sbjct 144 RRHADGSFSDEMNTVLDTL#ATRDFINWLLQTK 175 |
| Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRN 84 |||||||+|||||||+|#||||||||||+|||+ Sbjct 1 HSQGTFTNDYSKYLDTR#RAQDFVQWLMSTKRS 32 Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWL 78 |+ ||+||| | || +# |+ || || Sbjct 45 HADGTYTSDVSSYLQDQ#AAKKFVTWL 70 |
| Phodopus sungorus | Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTK 82 +||+ |+|+ + + ||| # +||+ ||+ || Sbjct 44 RRHADGSFSDEMNTILDSL#ATRDFINWLIQTK 75 Query 56 GTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKN 86 |||||| | ||+ +# |++|+ ||+ + ++ Sbjct 1 GTFTSDVSSYLEGQ#AAKEFIAWLVKGRGRRD 31 |
| Heloderma suspectum, venom, Peptide, 39 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMN 80 | +|||||| || ++ # + |++|| | Sbjct 1 HGEGTFTSDLSKQMEEE#AVRLFIEWLKN 28 |
| Bufo marinus | Query 52 RHSQGTFTSDYSKYLDSR#RAQDFVQWLM--NTKRNKNNIAKRHDEFERHA 99 ||++|||||| + +|+ +# |++|| ||+ ||| ++++ | ||| Sbjct 62 RHAEGTFTSDMTSFLEEK#AAKEFVDWLIKGRPKRNFSDVSAADDMDRRHA 111 Query 50 DKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNT 81 |+||+ |+||||++| || +# ||+|+ |++|| Sbjct 107 DRRHADGSFTSDFNKALDIK#AAQEFLDWIINT 138 Query 51 KRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDE 94 +||++||+|+| +++|+ +# |++|+ ||+ | ++ || | Sbjct 23 ERHAEGTYTNDVTQFLEEK#AAKEFIDWLLKGIPKKQRLS-RHAE 65 Query 73 DFVQWLMNTKRNKNNIAKRHDEFERHA 99 || |||||+||+ +++|+ +||||| Sbjct 1 DFAQWLMNSKRS-GGMSRRNVQFERHA 26 |
| Hydrolagus colliei | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 +|+| + + |||+ | |+||||||||+|#| +||||||++|||| | |+ +||| Sbjct 20 VSNPSTVTDVKRHTDGIFSSDYSKYLDNR#RTKDFVQWLLSTKRNGANT----DKTKRHA 74 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNN--IAKRHDEFER 97 ++ |||+ | +||| + | #+++ ||+ | | + +|+ ++||| | | Sbjct 68 DKTKRHADGIYTSDVASLTDYL#KSKRFVESLSNYNKRQNDRRMSKRHVEGAR 119 Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFE 96 | | ++ + +|+| ||+++# |+||+ ||+ + + + |+ | | Sbjct 116 EGARRTEEDYPNDFSSYLEAK#AAKDFIDWLIK-GQGRRDFAEESREIE 162 |
| Squalus acanthias | Query 44 PDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFE 96 |++| |+||+ |||||+ + ||+ # |++|+ |++|+| | | | | Sbjct 118 PEEM--DRRHADGTFTSEINIVLDTI#AAKEFLNWILNSK----TIQSRDSEIE 164 Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWL--MNTKRNKNNIAKRHDEFERH 98 ++ |||++||+||| | #+|+ || | | ++| +|+||| | || Sbjct 20 DKTKRHAEGTYTSDVDSLSDYF#KAKRFVDSLTSYNKRQNGRSISKRHVESTRH 72 Query 67 DSR#RAQDFVQWLMNTKRNKNNIAKRHDE 94 |+|#||+|||||||+|||| + ||| | Sbjct 1 DNR#RAKDFVQWLMSTKRNGDK-TKRHAE 27 Query 49 EDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLM 79 | ||++|++ | | ||+++# |+||+ ||+ Sbjct 68 ESTRHTEGSYR-DISSYLEAK#AAKDFINWLI 97 |
| Capra hircus | Query 48 NEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLM 79 +| +||++|||||| | ||+ +# |++|+ ||+ Sbjct 8 DEFERHAEGTFTSDVSSYLEGQ#AAKEFIAWLV 39 Query 86 NNIAKRHDEFERHA 99 |||||||||||||| Query 86 NNIAKRHDEFERHA 99 |
| Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | Query 47 MNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 ++ + ||+ | ||||||+ | # |+ +++ |+ || ++|++ +||+ Sbjct 75 VSRNARHADGVFTSDYSRLLGQI#SAKKYLESLIG-KRISSSISEDPVPIKRHS 126 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | Query 56 GTFTSDYSKYLDSR#RAQDFVQWL 78 ||+||| | || +# |++|| || Sbjct 3 GTYTSDVSSYLQDQ#AAKEFVSWL 25 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | Query 53 HSQGTFTSDYSKYLDSR#RAQDFVQWLMN 80 |+ || ||| | +|+ +# ++|+ ||++ Sbjct 1 HADGTLTSDISSFLEKQ#ATKEFIAWLVS 28 |
| Arvicanthis ansorgei | Query 47 MNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAKRHDEFERHA 99 ++ + ||+ | ||||||+ | # |+ +++ |+ || ++| + +||+ Sbjct 58 VSRNARHADGVFTSDYSRLLGQI#SAKKYLESLIG-KRISSSILEDPVPIKRHS 109 |
| Candida glabrata | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLDSR#RAQDFVQWLMNTKRNKNNIAK 90 |+| | ||+ | | | | + +|+ # || |+ + | +| +| Sbjct 351 LADGDDDEEDEEDSSSTSESSYDEEIDNE#EKQDIVEEEEDADENDDNFSK 400 |
[Site 3] SRRAQDFVQW77-LMNTKRNKNN
Trp77  Leu
Leu
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ser68 | Arg69 | Arg70 | Ala71 | Gln72 | Asp73 | Phe74 | Val75 | Gln76 | Trp77 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Leu78 | Met79 | Asn80 | Thr81 | Lys82 | Arg83 | Asn84 | Lys85 | Asn86 | Asn87 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| NEDKRHSQGTFTSDYSKYLDSRRAQDFVQWLMNTKRNKNNIAKRHDEFERHAEGTFTSDV |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 126.00 | 10 | glucagon, isoform CRA_b |
| 2 | N/A | 80.90 | 45 | 0 |
| 3 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | 64.70 | 1 | AAB31092.1 |
| 4 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | 61.60 | 1 | glucagon=proglucagon-derived 3.5 kda peptide |
| 5 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | 60.80 | 1 | glucagon II |
| 6 | Contains: Glucagon; Glucagon-like peptide | 60.10 | 4 | GLUC_AMICA Glucagon precursor |
| 7 | Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | 60.10 | 1 | glucagon |
| 8 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | 58.50 | 1 | glucagon G-29 |
| 9 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | 57.80 | 1 | GLUC1_ORENI Glucagon-1 (Glucagon I) gb |
| 10 | Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | 56.20 | 1 | GLUC_LAMFL Glucagon gb |
| 11 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | 53.50 | 1 | glucagon |
| 12 | deshis1, Desphe6, Glu9 | 53.50 | 1 | A Chain A, Nmr Solution Structure Of The Glucagon |
| 13 | Monodelphis domestica | 50.10 | 3 | PREDICTED: similar to Gastric inhibitory polypepti |
| 14 | Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | 48.50 | 1 | GLUC1_PETMA Glucagon-1 precursor |
| 15 | Hoplobatrachus rugulosus | 46.60 | 1 | AF324209_1 proglucagon |
| 16 | Bufo marinus | 46.60 | 1 | proglucagon |
| 17 | Gallus gallus | 46.20 | 4 | glucagon isoform 1 |
| 18 | Meleagris gallopavo | 46.20 | 2 | preproglucagon B |
| 19 | Xenopus laevis | 45.80 | 5 | proglucagon II |
| 20 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | 44.70 | 1 | GLUC_HELSU Glucagon precursor |
| 21 | Rattus norvegicus | 43.90 | 6 | Glucose-dependent insulinotropic peptide |
| 22 | Canis familiaris | 43.90 | 2 | PREDICTED: similar to Gastric inhibitory polypepti |
| 23 | Ictalurus punctatus | 43.10 | 3 | proglucagon I |
| 24 | Ambloplites rupestris | 43.10 | 1 | proglucagon |
| 25 | Xenopus tropicalis | 42.70 | 2 | glucagon |
| 26 | Capra hircus | 42.70 | 1 | glucagon |
| 27 | Bos taurus | 42.40 | 5 | vasoactive intestinal peptide |
| 28 | Oncorhynchus tshawytscha | 42.40 | 1 | S78474_1 proglucagon |
| 29 | Danio rerio | 42.00 | 6 | hypothetical protein LOC494052 |
| 30 | Macaca mulatta | 42.00 | 3 | PREDICTED: vasoactive intestinal peptide |
| 31 | Pan troglodytes | 42.00 | 2 | PREDICTED: gastric inhibitory polypeptide |
| 32 | Squalus acanthias | 41.60 | 1 | proglucagon |
| 33 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | 41.20 | 1 | GLUC2_LOPAM Glucagon-2 precursor (Glucagon II) |
| 34 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | 41.20 | 1 | glucagon-like peptide, GLP |
| 35 | Mus musculus | 40.80 | 6 | gastric inhibitory polypeptide |
| 36 | Sebastes caurinus | 40.80 | 4 | proglucagon I |
| 37 | Heloderma suspectum | 40.40 | 2 | proglucagon |
| 38 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | 40.00 | 2 | GLUC_CARAU Glucagon precursor |
| 39 | Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC2_ONCMY Glucagon-2 precursor (Glucagon II) |
| 40 | Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC1_ONCMY Glucagon-1 precursor (Glucagon I) |
| 41 | Heloderma horridum | 40.00 | 1 | EXE3_HELHO Exendin-3 precursor emb |
| 42 | Tetraodon nigroviridis | 39.30 | 3 | unnamed protein product |
| 43 | Protopterus dolloi | 38.90 | 1 | proglucagon |
| 44 | Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | 38.90 | 1 | VIP_MOUSE VIP peptides precursor |
| 45 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | 38.90 | 1 | glucagon-like peptide-2, GLP-2=proglucagon-derived |
| 46 | Agkistrodon piscivorus | 38.50 | 1 | proglucagon |
| 47 | Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | 38.10 | 1 | GLUC2_PETMA Glucagon-2 precursor |
| 48 | Arvicanthis ansorgei | 38.10 | 1 | vasoactive intestinal polypeptide |
| 49 | synthetic construct | 37.00 | 1 | Homo sapiens glucagon |
| 50 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | 36.60 | 1 | GLUC1_LOPAM Glucagon-1 precursor (Glucagon I) |
| 51 | Phodopus sungorus | 36.60 | 1 | glucagon precursor |
| 52 | Sus scrofa | 36.20 | 1 | glucagon |
| 53 | Neoceratodus forsteri | 36.20 | 1 | proglucagon |
| 54 | Canis lupus familiaris | 35.80 | 1 | glucagon |
| 55 | Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | 35.40 | 1 | GLUC_LEPSP Glucagon precursor |
| 56 | Heloderma suspectum, venom, Peptide, 39 aa | 35.40 | 1 | A Chain A, Solution Structure Of Exendin-4 In 30-V |
| 57 | Hydrolagus colliei | 34.30 | 1 | proglucagon |
| 58 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | 33.50 | 1 | GLUC2_ORENI Glucagon-2 (Glucagon II) gb |
| 59 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | 33.50 | 1 | glucagon-like peptide-1, GLP-1=proglucagon-derived |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSD 106 ||||||||||||||||||||||||||||||#|||||||+||||||||||||||||||||| Sbjct 131 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSD 189 |
| N/A | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIA 89 |||||||||||||||||||||||||#|||||||||||| Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIA 89 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRN 84 ||+||||||||||+|+|||+|||||#||+|||| Sbjct 1 HSEGTFTSDYSKYMDNRRAKDFVQW#LMSTKRN 32 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 |||||||||||||||+||||||+||#||+| Sbjct 1 HSQGTFTSDYSKYLDNRRAQDFIQW#LMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRN 84 ||+|||++||||||++|||||||||#| |+||| Sbjct 1 HSEGTFSNDYSKYLETRRAQDFVQW#LKNSKRN 32 |
| Contains: Glucagon; Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 |||||||+|||||+|+|||||||||#||+| Sbjct 1 HSQGTFTNDYSKYMDTRRAQDFVQW#LMST 29 |
| Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 |||||||+|||||+|+|||||||||#||+| Sbjct 1 HSQGTFTNDYSKYMDTRRAQDFVQW#LMST 29 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 ||+||||||||||+|+|||+|||||#||+| Sbjct 1 HSEGTFTSDYSKYMDNRRAKDFVQW#LMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRN 84 ||+|||++||||||+ |+|||||+|#||| ||+ Sbjct 1 HSEGTFSNDYSKYLEDRKAQDFVRW#LMNNKRS 32 |
| Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 ||||+|||||||||||++|+||| |#|||| Sbjct 1 HSQGSFTSDYSKYLDSKQAKDFVIW#LMNT 29 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMN 80 ||+|||||||||||++++|+|||+|#||| Sbjct 1 HSEGTFTSDYSKYLENKQAKDFVRW#LMN 28 |
| deshis1, Desphe6, Glu9 | Query 54 SQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 |||| ||+||||||||||||||||#|||| Sbjct 1 SQGT-TSEYSKYLDSRRAQDFVQW#LMNT 27 |
| Monodelphis domestica | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGT 102 +|+++||| |||| +| |||| |#|++ | || + ||+ || |+|| Sbjct 51 QRYAEGTFISDYSITMDKIMQQDFVNW#LLSQKGKKN--SWRHNITERKADGT 100 |
| Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||||||||||++++|+|||+|#||| || + + + |||+||||+|+ Sbjct 41 KRHSEGTFTSDYSKYLENKQAKDFVRW#LMNAKRGGSELQR------RHADGTFTNDM 91 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LM---NTKRNKNNIAKRHDEFERHAE 100 +||+ ||||+| + |||++ |+||| |#| ++|+ + + | +|||| Sbjct 80 RRHADGTFTNDMTSYLDAKAARDFVSW#LARSDKSRRDGGDHLAENSEDKRHAE 132 |
| Hoplobatrachus rugulosus | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 || |||||||||||||||||||||||||||#|||+||+ |+||+ +||||||||+|+|| Sbjct 48 NEVKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNSKRS-GGISKRNVQFERHAEGTYTNDV 106 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 +||++||+|+| +++|+ + |++|+ |#|+ | | ++ ||||||||||+ Sbjct 95 ERHAEGTYTNDVTQFLEEKAAKEFIDW#LIKGKPKKQRLS-------RHAEGTFTSDM 144 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LM--NTKRNKNNIAKRHDEFERHAEGTFTSD 106 ||++|||||| + ||+ + |++|| |#|+ ||| +|+ + |||+|+|||| Sbjct 134 RHAEGTFTSDMTSYLEEKAAKEFVDW#LIKGRPKRNFPDISAEEMD-RRHADGSFTSD 189 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 |+||+ |+||||++| || + ||+|+ |#++|| Sbjct 178 DRRHADGSFTSDFNKALDIKAAQEFLDW#IINT 209 |
| Bufo marinus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LM--NTKRNKNNIAKRHDEFERHAEGTFTSD 106 ||++|||||| + +|+ + |++|| |#|+ ||| ++++ | |||+|+|||| Sbjct 62 RHAEGTFTSDMTSFLEEKAAKEFVDW#LIKGRPKRNFSDVSAADDMDRRHADGSFTSD 118 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 +||++||+|+| +++|+ + |++|+ |#|+ | ++ ||||||||||+ Sbjct 23 ERHAEGTYTNDVTQFLEEKAAKEFIDW#LLKGIPKKQRLS-------RHAEGTFTSDM 72 Query 73 DFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 || ||#|||+||+ +++|+ +||||||||+|+|| Sbjct 1 DFAQW#LMNSKRS-GGMSRRNVQFERHAEGTYTNDV 34 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNT 81 |+||+ |+||||++| || + ||+|+ |#++|| Sbjct 107 DRRHADGSFTSDFNKALDIKAAQEFLDW#IINT 138 |
| Gallus gallus | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRN------------------KNNIA 89 || |||||||||||||||||||||||||||#||+|||| | |+ Sbjct 50 NEVKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMSTKRNGQQGQEDKENDKFPDQLSSNAIS 109 Query 90 KRHDEFERHAEGTFTSDV 107 ||| |||||||||+|||+ Sbjct 110 KRHSEFERHAEGTYTSDI 127 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAK---RHDEFERHAEGTFT 104 +| +||++||+||| + ||+ + |++|+ |#|+| + ++ | + |||+|||| Sbjct 113 SEFERHAEGTYTSDITSYLEGQAAKEFIAW#LVNGRGRRDFPEKALMAEEMGRRHADGTFT 172 Query 105 SDV 107 ||+ Sbjct 173 SDI 175 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNI 88 +||+ |||||| +| || |++|++|#|+||| + ++ Sbjct 164 RRHADGTFTSDINKILDDMAAKEFLKW#LINTKVTQRDL 201 |
| Meleagris gallopavo | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRN------------------KNNIA 89 || |||||||||||||||||||||||||||#||+|||| | |+ Sbjct 50 NEVKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMSTKRNGQQGQEDKENDEFPDQLSSNAIS 109 Query 90 KRHDEFERHAEGTFTSDV 107 ||| |||||||||+|||+ Sbjct 110 KRHSEFERHAEGTYTSDI 127 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAK---RHDEFERHAEGTFT 104 +| +||++||+||| + ||+ + |++|+ |#|+| + ++ | + |||+|||| Sbjct 113 SEFERHAEGTYTSDITSYLEGQAAKEFIAW#LVNGRGRRDFPEKALMAEEMGRRHADGTFT 172 Query 105 SDV 107 ||+ Sbjct 173 SDI 175 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNI 88 +||+ |||||| +| || |++|++|#|+||| + ++ Sbjct 164 RRHADGTFTSDINKILDDMAAKEFLKW#LINTKVTQRDL 201 |
| Xenopus laevis | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 | |||||||||||||||||||||||||||#||||||+ +++|+ ++|||||||||||| Sbjct 49 EVKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRS-GGLSRRNADYERHAEGTFTSDV 106 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 | +||++||+|+| ++||+ + + |++|#|+ | | + ||||||||+|+ Sbjct 138 EIERHAEGTYTNDVTEYLEEKATKAFIEW#LIKGKPKK-------IRYSRHAEGTFTNDM 189 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRH 92 ||++||||+| + ||+ + |++|| |#|+| + + ++ + | Sbjct 179 RHAEGTFTNDMTNYLEEKAAKEFVGW#LINGRPKRKDLLEEH 219 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKR------------------NKNNIA 89 || |||||||||||||||||+|||||||||#|||||| + | +| Sbjct 48 NEVKRHSQGTFTSDYSKYLDTRRAQDFVQW#LMNTKRSGQQGVEEREKENLLDQLSSNGLA 107 Query 90 KRHDEFERHAEGTFTSDV 107 + | |+||||+| +|||+ Sbjct 108 RHHAEYERHADGRYTSDI 125 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAK---RHDEFERHAEGTFTS 105 | +||+ | +||| | ||+ + |++|+ |#|+| + ++ + + | |||+||||| Sbjct 112 EYERHADGRYTSDISSYLEGQAAKEFIAW#LVNGRGRRDFLEEAGTADDIGRRHADGTFTS 171 Query 106 D 106 | Query 106 D 106 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNI 88 +||+ |||||||++ || |+|++|#|+| | + ++ Sbjct 162 RRHADGTFTSDYNQLLDDIATQEFLKW#LINQKVTQRDL 199 |
| Rattus norvegicus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| | |||| |#|+ | ||+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDFVNW#LLAQKGKKND--WKHNLTQREA 88 |
| Canis familiaris | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| | |||| |#|+ | ||+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDFVNW#LLAQKGKKND--WKHNITQREA 88 |
| Ictalurus punctatus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 +|||+|||++||||||++|||||||||#|||+||+ + |||+||+|||| Sbjct 49 RRHSEGTFSNDYSKYLETRRAQDFVQW#LMNSKRSDS-------PARRHADGTYTSDV 98 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEF--ERHAEGTFTSDV 107 +||+ ||+||| | || + |+||+ |#| + + + + | + || +|+||||| Sbjct 87 RRHADGTYTSDVSSYLQDQAAKDFITW#LKSGQPKPDAVETRSVDTLRRRHVDGSFTSDV 145 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRH 92 +|| |+|||| +| ||| |++++||#+||+ + + ||| Sbjct 134 RRHVDGSFTSDVNKVLDSLAAKEYLQW#VMNSPSSAS--GKRH 173 |
| Ambloplites rupestris | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSD 106 |||||||+||+ ||+ |+||||++|#||| || + |++ |||+|||| | Sbjct 1 HSQGTFTNDYTNYLEDRQAQDFIRW#LMNNKR--SGAAEK-----RHADGTFTDD 47 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK-RNKNNIAKRHDEF-ERHAEGTFTSDV 107 +|||+ |||| | | + +||| #| + + | + +| + | || ||+||||+ Sbjct 36 EKRHADGTFTDDASSDFYDQAIKDFVAK#LKSGQDRREPETDRRREAFSRRHVEGSFTSDI 95 |
| Xenopus tropicalis | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 | ||||||||||||||||||||||||+||#||||||+ +++|+ ++|||||||||||| Sbjct 49 EVKRHSQGTFTSDYSKYLDSRRAQDFIQW#LMNTKRS-GGLSRRNADYERHAEGTFTSDV 106 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 +||++||+|+| ++||+ + |++|++|#|+| | | + ||||||||+|+ Sbjct 140 ERHAEGTYTNDVTEYLEEKAAKEFIEW#LINGKPKK-------IRYSRHAEGTFTNDM 189 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFE---RHAEGTFTSDV 107 ||++||||+| + ||+ + |++|| |#|+ | | | ++ | | |||+|+||+|+ Sbjct 179 RHAEGTFTNDMTNYLEEKAAKEFVGW#LIK-GRPKRNFSEAHSVEEMDRRHADGSFTNDI 236 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNI 88 |+||+ |+||+| +| || ||+|+ |#++||+ + ++ Sbjct 224 DRRHADGSFTNDINKVLDIIAAQEFLDW#VINTQVTERDL 262 |
| Capra hircus | Query 86 NNIAKRHDEFERHAEGTFTSDV 107 |||||||||||||||||||||| Query 86 NNIAKRHDEFERHAEGTFTSDV 107 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LM 79 +| +||++|||||| | ||+ + |++|+ |#|+ Sbjct 8 DEFERHAEGTFTSDVSSYLEGQAAKEFIAW#LV 39 |
| Bos taurus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSD 106 ||+ | ||||||+ | |+ +++ #|+ || |+|++ +||++ || + Sbjct 80 RHADGVFTSDYSRLLGQLSAKKYLES#LIG-KRVSNSISEDQGPIKRHSDAVFTDN 133 |
| Oncorhynchus tshawytscha | Query 54 SQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 | |||++||||| + | |||||||#|||+||+ +|||+||+|||| Sbjct 2 SXGTFSNDYSKYQEERMAQDFVQW#LMNSKRS-------GAPSKRHADGTYTSDV 48 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNK 85 |||+ ||+||| | || + |+||| |#| + + + Sbjct 37 KRHADGTYTSDVSTYLQDQAAKDFVSW#LKSGRARR 71 |
| Danio rerio | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++||||||++|||||||||#||| ||| ++ +||||||+|||| Sbjct 48 KRHSEGTFSNDYSKYLETRRAQDFVQW#LMNAKRNGGSV-------KRHAEGTYTSDV 97 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#L 78 |||++||+||| | || + || || |#| Sbjct 86 KRHAEGTYTSDVSSYLQDQAAQSFVAW#L 113 |
| Macaca mulatta | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSD 106 + + ||+ | ||||+|| | |+ +++ #|| || +||++ +||++ || + Sbjct 76 SRNARHADGVFTSDFSKLLGQLSAKKYLES#LMG-KRVSSNISEDPVPVKRHSDAVFTDN 133 |
| Pan troglodytes | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| |||| |#|+ | ||+ +|+ +| | Sbjct 51 RYAEGTFISDYSIAMDKIHQQDFVNW#LLAQKGKKND--WKHNITQREA 96 |
| Squalus acanthias | Query 67 DSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 |+|||+|||||#||+|||| |+ +||||||+|||| Sbjct 1 DNRRAKDFVQW#LMSTKRN-------GDKTKRHAEGTYTSDV 34 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#L--MNTKRNKNNIAKRHDEFERHAEGTF 103 ++ |||++||+||| | +|+ || #| | ++| +|+||| | || ||++ Sbjct 20 DKTKRHAEGTYTSDVDSLSDYFKAKRFVDS#LTSYNKRQNGRSISKRHVESTRHTEGSY 77 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKN---------NIAKRHDEFERHA 99 | ||++|++ | | ||+++ |+||+ |#|+ + + | + ||| Sbjct 68 ESTRHTEGSYR-DISSYLEAKAAKDFINW#LIKGRGRREFPEESKEIVNEVIPEEMDRRHA 126 Query 100 EGTFTSDV 107 +|||||++ Sbjct 127 DGTFTSEI 134 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFE 96 |+||+ |||||+ + ||+ |++|+ |#++|+| | | | | Sbjct 122 DRRHADGTFTSEINIVLDTIAAKEFLNW#ILNSK----TIQSRDSEIE 164 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++||||||++|||||||||#| |+||| | |||+||+|||| Sbjct 50 KRHSEGTFSNDYSKYLETRRAQDFVQW#LKNSKRN--------GLFRRHADGTYTSDV 98 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#L 78 +||+ ||+||| | || + |+||| |#| Sbjct 87 RRHADGTYTSDVSSYLQDQAAKDFVSW#L 114 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRN 84 |+ ||||+| + |||++ |+||| |#| + ++ Sbjct 1 HADGTFTNDMTSYLDAKAARDFVSW#LARSDKS 32 |
| Mus musculus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| | |||| |#|+ + |++ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDFVNW#LLAQRGKKSD--WKHNITQREA 88 |
| Sebastes caurinus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++|||+||+ |+|||||+|#||| ||+ || +|||+||||||| Sbjct 48 KRHSEGTFSNDYSRYLEERKAQDFVRW#LMNNKRS------GADE-KRHADGTFTSDV 97 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK-RNKNNIAKRHDEF-ERHAEGTFTSD 106 ++|||+ |||||| | || + +||| #| + + | ++ | + | || +|+|||| Sbjct 84 DEKRHADGTFTSDVSSYLKDQAIKDFVNR#LKSGQVRRESETEWRGEAFSRRHVDGSFTSD 143 Query 107 V 107 | Query 107 V 107 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDE 94 +|| |+|||| +| ||| |++++ |#+| +| + + ++ |+ Sbjct 133 RRHVDGSFTSDVNKVLDSMAAKEYLLW#VMTSKPSGESKKRQEDQ 176 |
| Heloderma suspectum | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKR------------------NKNNIA 89 || |||||||||||||||||+|||||||||#|||||| + | +| Sbjct 48 NEVKRHSQGTFTSDYSKYLDTRRAQDFVQW#LMNTKRSGQQGVEEREKENLLDQLSSNGLA 107 Query 90 KRHDEFERHAEGTFTSDV 107 + | |+||||+| +|||+ Sbjct 108 RHHAEYERHADGRYTSDI 125 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMN 80 | +||+ | +||| | ||+ + |++|+ |#|+| Sbjct 112 EYERHADGRYTSDISSYLEGQAAKEFIAW#LVN 143 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++||||||++|||||||+|#|||+| | + +||||||+|||+ Sbjct 48 KRHSEGTFSNDYSKYLETRRAQDFVEW#LMNSKSNGGSA-------KRHAEGTYTSDI 97 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#L 78 |||++||+||| | +| + ||+|| |#| Sbjct 86 KRHAEGTYTSDISSFLRDQAAQNFVAW#L 113 |
| Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 || |+|||++ |||| + | |+||+ |#|||+||+ +|||+||+|||| Sbjct 50 KRQSEGTFSNYYSKYQEERMARDFLHW#LMNSKRS-------GAPSKRHADGTYTSDV 99 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMN--TKRNKNNIAKRHDEFERHAEGTFTSDV 107 |||+ ||+||| | || + |+||| |#| + +| + || +|+||||| Sbjct 88 KRHADGTYTSDVSTYLQDQAAKDFVSW#LKSGPARRESAEESMNGPMSRRHVDGSFTSDV 146 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHD 93 +|| |+|||| +| ||| |++++ |#+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSLAAKEYLLW#VMTSKTSGKSNKRQED 177 |
| Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++||||| + | |||||||#|||+||+ +|||+||+|||| Sbjct 50 KRHSEGTFSNDYSKYQEERMAQDFVQW#LMNSKRS-------GAPSKRHADGTYTSDV 99 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMN--TKRNKNNIAKRHDEFERHAEGTFTSDV 107 |||+ ||+||| | || + |+||| |#| + +| ++ || +|+||||| Sbjct 88 KRHADGTYTSDVSTYLQDQAAKDFVSW#LKSGRARRESAEESRNGPMSRRHVDGSFTSDV 146 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHD 93 +|| |+|||| +| ||| |++++ |#+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSLAAKEYLLW#VMTSKTSGKSNKRQED 177 |
| Heloderma horridum | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMN 80 |||| |||||| || ++ + |++|#| | Sbjct 46 KRHSDGTFTSDLSKQMEEEAVRLFIEW#LKN 75 |
| Tetraodon nigroviridis | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++||||||++|||||||||#| |+||| + | |||+||+|||| Sbjct 787 KRHSEGTFSNDYSKYLETRRAQDFVQW#LKNSKRNGS-------LFRRHADGTYTSDV 836 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#L 78 +||+ ||+||| | || + |++|| |#| Sbjct 825 RRHADGTYTSDVSAYLQDQAAKEFVSW#L 852 |
| Protopterus dolloi | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 | |||||||| |||+| |+++ | | || #||+||| ++|||+| |||||||+|||+ Sbjct 28 EAKRHSQGTFMSDYAKLLEAKHALDLVQR#LMSTKRT-GAVSKRHNEIERHAEGTYTSDI 85 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFE-RHAEGTFTSD 106 || +||++||+||| | ||+ + +|++|#|| + ++ +| + |||+|| ||| Sbjct 71 NEIERHAEGTYTSDISSYLEGQAVNEFIRW#LMKGRGRRDFSETDIEEMDRRHADGTVTSD 130 Query 107 V 107 + Sbjct 131 I 131 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 |+||+ || ||| + |+| ++|+ |#|+| | Sbjct 119 DRRHADGTVTSDINSVLESIATREFLNW#LINAK 151 |
| Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSD 106 + + ||+ | ||||||+ | |+ +++ #|+ || ++|++ +||++ || + Sbjct 76 SRNARHADGVFTSDYSRLLGQISAKKYLES#LIG-KRISSSISEDPVPIKRHSDAVFTDN 133 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 |+ |+|||| +| ||+ |++|+ |#|++|| Sbjct 1 HADGSFTSDINKVLDTIAAKEFLNW#LISTK 30 |
| Agkistrodon piscivorus | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRH---DEFERHAEGTFTS 105 | +||+ ||+||| | ||+ + |++|+ |#|+| + ++ | | |||+||||| Sbjct 32 EYERHADGTYTSDISSYLEGQAAKEFIAW#LVNGRGRRDFSEGAHTGEDLGRRHADGTFTS 91 Query 106 D 106 | Query 106 D 106 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNI 88 +||+ |||||||+| || |+|++|#|+| | + ++ Sbjct 82 RRHADGTFTSDYNKLLDDIATQEFLKW#LINQKVTQRDL 119 Query 66 LDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 |+ | ++|+ #| + | +|+ | |+||||+||+|||+ Sbjct 9 LEDREKENFLDQ#LAS-----NGLARPHAEYERHADGTYTSDI 45 |
| Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 |||||+|||||||+|| ++|+||| |#|+|||| + + +||++|+||+|+ Sbjct 43 RHSQGSFTSDYSKHLDVKQAKDFVTW#LLNTKRRGVDAQAGANLEKRHSDGSFTNDM 98 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKR 83 | +|||| |+||+| + || |++|++|#| | Sbjct 84 NLEKRHSDGSFTNDMNVMLDRMSAKNFLEW#LKQQGR 119 |
| Arvicanthis ansorgei | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSD 106 + + ||+ | ||||||+ | |+ +++ #|+ || ++| + +||++ || + Sbjct 59 SRNARHADGVFTSDYSRLLGQISAKKYLES#LIG-KRISSSILEDPVPIKRHSDAVFTDN 116 |
| synthetic construct | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 ||||||||||||||||||||||||||||||#|||||||+|||||||||||||||||||||| Sbjct 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNRNNIAKRHDEFERHAEGTFTSDV 107 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKN---NIAKRHDEFERHAEGTFT 104 +| +||++|||||| | ||+ + |++|+ |#|+ + ++ +| + |||+|+|+ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQAAKEFIAW#LVKGRGRRDFPEEVAIVEELGRRHADGSFS 152 Query 105 SDV 107 ++ Sbjct 153 DEM 155 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 +||+ |+|+ + + ||+ |+||+ |#|+ || Sbjct 144 RRHADGSFSDEMNTILDNLAARDFINW#LIQTK 175 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 ||||+|||++||||||+ |+||+||+|#||| || + +|++ |||+||||||| Sbjct 51 KRHSEGTFSNDYSKYLEDRKAQEFVRW#LMNNKR--SGVAEK-----RHADGTFTSDV 100 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQW#L 78 +|||+ |||||| | || + +||| #| Sbjct 88 EKRHADGTFTSDVSSYLKDQAIKDFVDR#L 116 |
| Phodopus sungorus | Query 56 GTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKN---NIAKRHDEFERHAEGTFTSDV 107 |||||| | ||+ + |++|+ |#|+ + ++ + + |||+|+|+ ++ Sbjct 1 GTFTSDVSSYLEGQAAKEFIAW#LVKGRGRRDFPEEVTIVEELGRRHADGSFSDEM 55 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 +||+ |+|+ + + ||| +||+ |#|+ || Sbjct 44 RRHADGSFSDEMNTILDSLATRDFINW#LIQTK 75 |
| Sus scrofa | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 |||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKN---NIAKRHDEFERHAEGTFT 104 +| +||++|||||| | ||+ + |++|+ |#|+ + ++ + + |||+|+|+ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQAAKEFIAW#LVKGRGRRDFPEEVTIVEELRRRHADGSFS 152 Query 105 SDV 107 ++ Sbjct 153 DEM 155 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 +||+ |+|+ + + ||+ +||+ |#|++|| Sbjct 144 RRHADGSFSDEMNTVLDNLATRDFINW#LLHTK 175 |
| Neoceratodus forsteri | Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 | |||||||| |||+| |++| | |||| #||||||| ++||| | |||||||+|||+ Sbjct 20 EAKRHSQGTFMSDYAKLLEARHALDFVQR#LMNTKRN-GGVSKRHSEIERHAEGTYTSDI 77 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEF-ERHAEGTFTSD 106 +| +||++||+||| | ||+ + | +||+|#|+ + ++ +| |||+|| |+| Sbjct 63 SEIERHAEGTYTSDISSYLEGQAANEFVRW#LLKGRGRRDFSDTDAEEMGRRHADGTITND 122 Query 107 V 107 + Sbjct 123 M 123 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 +||+ || |+| + |+| ++|+ |#|+|+| Sbjct 112 RRHADGTITNDMNNVLESIATREFLNW#LINSK 143 |
| Canis lupus familiaris | Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKN---NIAKRHDEFERHAEGTFT 104 +| +||++|||||| | ||+ + |++|+ |#|+ + ++ +| + |||+|+|+ Sbjct 93 DEFERHAEGTFTSDVSSYLEGQAAKEFIAW#LVKGRGRRDFPEEVAIVEEFRRRHADGSFS 152 Query 105 SDV 107 ++ Sbjct 153 DEM 155 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTK 82 +||+ |+|+ + + ||+ +||+ |#|+ || Sbjct 144 RRHADGSFSDEMNTVLDTLATRDFINW#LLQTK 175 |
| Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 |||||||+|||||||+|||||||||#||+|||+ | ||+||+|||| Sbjct 1 HSQGTFTNDYSKYLDTRRAQDFVQW#LMSTKRS-GGITXXXXXXXXHADGTYTSDV 54 Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#L 78 |+ ||+||| | || + |+ || |#| Sbjct 45 HADGTYTSDVSSYLQDQAAKKFVTW#L 70 |
| Heloderma suspectum, venom, Peptide, 39 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMN 80 | +|||||| || ++ + |++|#| | Sbjct 1 HGEGTFTSDLSKQMEEEAVRLFIEW#LKN 28 |
| Hydrolagus colliei | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFERHAEGTFTSDV 107 |||+ | |+||||||||+|| +|||||#|++|||| | |+ +|||+| +|||| Sbjct 30 KRHTDGIFSSDYSKYLDNRRTKDFVQW#LLSTKRNGANT----DKTKRHADGIYTSDV 82 Query 48 NEDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNN--IAKRHDEFERHAEGTFTS 105 ++ |||+ | +||| + | +++ ||+ #| | + +|+ ++||| | | | + + Sbjct 68 DKTKRHADGIYTSDVASLTDYLKSKRFVES#LSNYNKRQNDRRMSKRHVEGARRTEEDYPN 127 Query 106 D 106 | Query 106 D 106 Query 49 EDKRHSQGTFTSDYSKYLDSRRAQDFVQW#LMNTKRNKNNIAKRHDEFE 96 | | ++ + +|+| ||+++ |+||+ |#|+ + + + |+ | | Sbjct 116 EGARRTEEDYPNDFSSYLEAKAAKDFIDW#LIK-GQGRRDFAEESREIE 162 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | Query 56 GTFTSDYSKYLDSRRAQDFVQW#L 78 ||+||| | || + |++|| |#| Sbjct 3 GTYTSDVSSYLQDQAAKEFVSW#L 25 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQW#LMN 80 |+ || ||| | +|+ + ++|+ |#|++ Sbjct 1 HADGTLTSDISSFLEKQATKEFIAW#LVS 28 |
[Site 4] FTSDYSKYLD67-SRRAQDFVQW
Asp67  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Phe58 | Thr59 | Ser60 | Asp61 | Tyr62 | Ser63 | Lys64 | Tyr65 | Leu66 | Asp67 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser68 | Arg69 | Arg70 | Ala71 | Gln72 | Asp73 | Phe74 | Val75 | Gln76 | Trp77 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| TDPLSDPDQMNEDKRHSQGTFTSDYSKYLDSRRAQDFVQWLMNTKRNKNNIAKRHDEFER |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 126.00 | 9 | glucagon, isoform CRA_b |
| 2 | Pan troglodytes | 105.00 | 2 | PREDICTED: fibroblast activation protein, alpha su |
| 3 | N/A | 80.90 | 43 | 0 |
| 4 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | 64.70 | 1 | AAB31092.1 |
| 5 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | 61.60 | 1 | glucagon=proglucagon-derived 3.5 kda peptide |
| 6 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | 60.80 | 1 | glucagon II |
| 7 | Contains: Glucagon; Glucagon-like peptide | 60.10 | 4 | GLUC_AMICA Glucagon precursor |
| 8 | Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | 60.10 | 1 | glucagon |
| 9 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | 58.50 | 1 | glucagon G-29 |
| 10 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | 57.80 | 1 | GLUC1_ORENI Glucagon-1 (Glucagon I) gb |
| 11 | Ambloplites rupestris | 57.40 | 1 | proglucagon |
| 12 | Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | 56.20 | 1 | GLUC_LAMFL Glucagon gb |
| 13 | deshis1, Desphe6, Glu9 | 53.50 | 1 | A Chain A, Nmr Solution Structure Of The Glucagon |
| 14 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | 53.50 | 1 | glucagon |
| 15 | Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | 45.80 | 1 | GLUC1_PETMA Glucagon-1 precursor |
| 16 | Monodelphis domestica | 43.90 | 3 | PREDICTED: similar to Gastric inhibitory polypepti |
| 17 | Rattus norvegicus | 43.50 | 3 | Glucose-dependent insulinotropic peptide |
| 18 | Canis familiaris | 43.50 | 2 | PREDICTED: similar to Gastric inhibitory polypepti |
| 19 | Gallus gallus | 43.10 | 4 | glucagon isoform 1 |
| 20 | Meleagris gallopavo | 43.10 | 2 | preproglucagon B |
| 21 | Agkistrodon piscivorus | 43.10 | 1 | proglucagon |
| 22 | Xenopus tropicalis | 42.70 | 2 | glucagon |
| 23 | Capra hircus | 42.70 | 1 | glucagon |
| 24 | Hoplobatrachus rugulosus | 42.70 | 1 | AF324209_1 proglucagon |
| 25 | Xenopus laevis | 42.40 | 5 | proglucagon I |
| 26 | Oncorhynchus tshawytscha | 42.40 | 1 | S78474_1 proglucagon |
| 27 | Danio rerio | 42.00 | 4 | hypothetical protein LOC494052 |
| 28 | Bos taurus | 41.60 | 5 | PREDICTED: similar to Gastric inhibitory polypepti |
| 29 | Macaca mulatta | 41.60 | 3 | PREDICTED: gastric inhibitory polypeptide |
| 30 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | 41.20 | 1 | glucagon-like peptide, GLP |
| 31 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | 41.20 | 1 | GLUC2_LOPAM Glucagon-2 precursor (Glucagon II) |
| 32 | Ictalurus punctatus | 40.80 | 3 | proglucagon I |
| 33 | Heloderma horridum | 40.80 | 1 | EXE3_HELHO Exendin-3 precursor emb |
| 34 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | 40.80 | 1 | GLUC_HELSU Glucagon precursor |
| 35 | Mus musculus | 40.40 | 5 | gastric inhibitory polypeptide |
| 36 | Heloderma suspectum | 40.40 | 2 | proglucagon |
| 37 | Protopterus dolloi | 40.40 | 1 | proglucagon |
| 38 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | 40.00 | 2 | GLUC_CARAU Glucagon precursor |
| 39 | Bufo marinus | 40.00 | 1 | proglucagon |
| 40 | Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC1_ONCMY Glucagon-1 precursor (Glucagon I) |
| 41 | Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC2_ONCMY Glucagon-2 precursor (Glucagon II) |
| 42 | Neoceratodus forsteri | 39.70 | 1 | proglucagon |
| 43 | Sebastes caurinus | 39.30 | 4 | proglucagon II |
| 44 | Tetraodon nigroviridis | 39.30 | 2 | unnamed protein product |
| 45 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | 38.90 | 1 | glucagon-like peptide-2, GLP-2=proglucagon-derived |
| 46 | Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | 38.10 | 1 | GLUC2_PETMA Glucagon-2 precursor |
| 47 | synthetic construct | 37.00 | 1 | Homo sapiens glucagon |
| 48 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | 36.60 | 1 | GLUC1_LOPAM Glucagon-1 precursor (Glucagon I) |
| 49 | Sus scrofa | 36.20 | 1 | glucagon |
| 50 | Canis lupus familiaris | 35.80 | 1 | glucagon |
| 51 | Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | 35.40 | 1 | GLUC_LEPSP Glucagon precursor |
| 52 | Heloderma suspectum, venom, Peptide, 39 aa | 35.40 | 1 | A Chain A, Solution Structure Of Exendin-4 In 30-V |
| 53 | Phodopus sungorus | 35.40 | 1 | glucagon precursor |
| 54 | Squalus acanthias | 34.30 | 1 | proglucagon |
| 55 | Hydrolagus colliei | 34.30 | 1 | proglucagon |
| 56 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | 33.50 | 1 | GLUC2_ORENI Glucagon-2 (Glucagon II) gb |
| 57 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | 33.50 | 1 | glucagon-like peptide-1, GLP-1=proglucagon-derived |
| 58 | Candida glabrata | 33.10 | 1 | unnamed protein product |
| 59 | swine, intestines, Peptide Partial, 56 aa | 32.30 | 1 | secretin precursor, N-prosecretin, secretin-(-9 to |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 |||||||||||||||||||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 122 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 180 |
| Pan troglodytes | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDE 94 |||||||||||||||||||||||||||||#||||||||||||||||| +++| || | Sbjct 961 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN-SSVAYRHAE 1015 |
| N/A | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIA 89 |||||||||||||||#|||||||||||||||||||||| Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIA 89 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN 84 ||+||||||||||+|#+|||+|||||||+|||| Sbjct 1 HSEGTFTSDYSKYMD#NRRAKDFVQWLMSTKRN 32 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 |||||||||||||||#+||||||+||||+| Sbjct 1 HSQGTFTSDYSKYLD#NRRAQDFIQWLMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN 84 ||+|||++||||||+#+|||||||||| |+||| Sbjct 1 HSEGTFSNDYSKYLE#TRRAQDFVQWLKNSKRN 32 |
| Contains: Glucagon; Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 |||||||+|||||+|#+|||||||||||+| Sbjct 1 HSQGTFTNDYSKYMD#TRRAQDFVQWLMST 29 |
| Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 |||||||+|||||+|#+|||||||||||+| Sbjct 1 HSQGTFTNDYSKYMD#TRRAQDFVQWLMST 29 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 ||+||||||||||+|#+|||+|||||||+| Sbjct 1 HSEGTFTSDYSKYMD#NRRAKDFVQWLMST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN 84 ||+|||++||||||+# |+|||||+|||| ||+ Sbjct 1 HSEGTFSNDYSKYLE#DRKAQDFVRWLMNNKRS 32 |
| Ambloplites rupestris | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRH 92 |||||||+||+ ||+# |+||||++|||| ||+ ||| Sbjct 1 HSQGTFTNDYTNYLE#DRQAQDFIRWLMNNKRS-GAAEKRH 39 |
| Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 ||||+||||||||||#|++|+||| ||||| Sbjct 1 HSQGSFTSDYSKYLD#SKQAKDFVIWLMNT 29 |
| deshis1, Desphe6, Glu9 | Query 54 SQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 |||| ||+||||||#|||||||||||||| Sbjct 1 SQGT-TSEYSKYLD#SRRAQDFVQWLMNT 27 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMN 80 ||+|||||||||||+#+++|+|||+|||| Sbjct 1 HSEGTFTSDYSKYLE#NKQAKDFVRWLMN 28 |
| Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRH 92 ||||+|||||||||||+#+++|+|||+|||| || + + +|| Sbjct 41 KRHSEGTFTSDYSKYLE#NKQAKDFVRWLMNAKRGGSELQRRH 82 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNK---------NNIAKRHDE 94 +||+ ||||+| + |||#++ |+||| || + +++ |+ ||| | Sbjct 80 RRHADGTFTNDMTSYLD#AKAARDFVSWLARSDKSRRDGGDHLAENSEDKRHAE 132 |
| Monodelphis domestica | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 +|+++||| |||| +|# |||| ||++ | || + ||+ || Sbjct 51 QRYAEGTFISDYSITMD#KIMQQDFVNWLLSQKGKKN--SWRHNITER 95 |
| Rattus norvegicus | Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNN 87 |+++||| |||| +|# | |||| ||+ | ||+ Sbjct 43 RYAEGTFISDYSIAMD#KIRQQDFVNWLLAQKGKKND 78 |
| Canis familiaris | Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNN 87 |+++||| |||| +|# | |||| ||+ | ||+ Sbjct 43 RYAEGTFISDYSIAMD#KIRQQDFVNWLLAQKGKKND 78 |
| Gallus gallus | Query 38 TDPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN------------- 84 ++|| + |+|| |||||||||||||||||#||||||||||||+|||| Sbjct 40 SEPLDESRQLNEVKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMSTKRNGQQGQEDKENDKF 99 Query 85 -----KNNIAKRHDEFER 97 | |+||| |||| Sbjct 100 PDQLSSNAISKRHSEFER 117 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNI 88 +||+ |||||| +| ||# |++|++||+||| + ++ Sbjct 164 RRHADGTFTSDINKILD#DMAAKEFLKWLINTKVTQRDL 201 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKN 86 +| +||++||+||| + ||+# + |++|+ ||+| + ++ Sbjct 113 SEFERHAEGTYTSDITSYLE#GQAAKEFIAWLVNGRGRRD 151 |
| Meleagris gallopavo | Query 38 TDPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN------------- 84 ++|| + |+|| |||||||||||||||||#||||||||||||+|||| Sbjct 40 SEPLDESRQLNEVKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMSTKRNGQQGQEDKENDEF 99 Query 85 -----KNNIAKRHDEFER 97 | |+||| |||| Sbjct 100 PDQLSSNAISKRHSEFER 117 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNI 88 +||+ |||||| +| ||# |++|++||+||| + ++ Sbjct 164 RRHADGTFTSDINKILD#DMAAKEFLKWLINTKVTQRDL 201 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKN 86 +| +||++||+||| + ||+# + |++|+ ||+| + ++ Sbjct 113 SEFERHAEGTYTSDITSYLE#GQAAKEFIAWLVNGRGRRD 151 |
| Agkistrodon piscivorus | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNI 88 +||+ |||||||+| ||# |+|++||+| | + ++ Sbjct 82 RRHADGTFTSDYNKLLD#DIATQEFLKWLINQKVTQRDL 119 Query 49 EDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKN 86 | +||+ ||+||| | ||+# + |++|+ ||+| + ++ Sbjct 32 EYERHADGTYTSDISSYLE#GQAAKEFIAWLVNGRGRRD 69 |
| Xenopus tropicalis | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 + + | |+ | |||||||||||||||||#|||||||+||||||||+ +++|+ ++|| Sbjct 39 EAVDDSQQLKEVKRHSQGTFTSDYSKYLD#SRRAQDFIQWLMNTKRS-GGLSRRNADYER 96 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 +||++|||||| +++||# + |++|+ ||+| +| |++|+ + || Sbjct 95 ERHAEGTFTSDVTQHLD#EKAAKEFIDWLINGGPSKEIISRRNADVER 141 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDE 94 +||++||+|+| ++||+# + |++|++||+| | | + || | Sbjct 140 ERHAEGTYTNDVTEYLE#EKAAKEFIEWLINGKPKKIRYS-RHAE 182 Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRH--DEFER 97 ||++||||+| + ||+# + |++|| ||+ | | | ++ | +| +| Sbjct 179 RHAEGTFTNDMTNYLE#EKAAKEFVGWLIK-GRPKRNFSEAHSVEEMDR 225 Query 50 DKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNI 88 |+||+ |+||+| +| ||# ||+|+ |++||+ + ++ Sbjct 224 DRRHADGSFTNDINKVLD#IIAAQEFLDWVINTQVTERDL 262 |
| Capra hircus | Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLM 79 +| +||++|||||| | ||+# + |++|+ ||+ Sbjct 8 DEFERHAEGTFTSDVSSYLE#GQAAKEFIAWLV 39 |
| Hoplobatrachus rugulosus | Query 38 TDPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 |+ | | | || |||||||||||||||||#|||||||||||||+||+ |+||+ +||| Sbjct 38 TEALDDSDHHNEVKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNSKRS-GGISKRNVQFER 96 Query 50 DKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 |+||+ |+||||++| ||# + ||+|+ |++|| Sbjct 178 DRRHADGSFTSDFNKALD#IKAAQEFLDWIINT 209 Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKN 86 ||++|||||| + ||+# + |++|| ||+ + +| Sbjct 134 RHAEGTFTSDMTSYLE#EKAAKEFVDWLIKGRPKRN 168 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDE 94 +||++||+|+| +++|+# + |++|+ ||+ | | ++ || | Sbjct 95 ERHAEGTYTNDVTQFLE#EKAAKEFIDWLIKGKPKKQRLS-RHAE 137 |
| Xenopus laevis | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 + + | +|+ | |||||||||||||||||#||||||||||||||||+ +++|+ ++|| Sbjct 39 EAVDDSEQLKEVKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRS-GELSRRNADYER 96 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 +||++|||||| ++ ||# + |++|+ ||+| +| |++|+ |||| Sbjct 95 ERHAEGTFTSDVTQQLD#EKAAKEFIDWLINGGPSKEIISRRNAEFER 141 Query 50 DKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNI 88 |+||+ |+||+| +| ||# ||+|+ |++||+ + ++ Sbjct 224 DRRHADGSFTNDINKVLD#IIAAQEFLDWVINTQETERDL 262 Query 49 EDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDE 94 | +||++||+|+| ++||+# + |++|++||+ | | + || | Sbjct 138 EFERHAEGTYTNDVTEYLE#EKAAKEFIEWLIKGKPKKIRYS-RHAE 182 Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRH--DEFER 97 ||++||||+| + ||+# + |++|| ||+ | | | ++ | +| +| Sbjct 179 RHAEGTFTNDMTNYLE#EKAAKEFVGWLIK-GRPKRNFSEVHSVEEMDR 225 |
| Oncorhynchus tshawytscha | Query 54 SQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRH 92 | |||++||||| +# | ||||||||||+||+ +||| Sbjct 2 SXGTFSNDYSKYQE#ERMAQDFVQWLMNSKRS-GAPSKRH 39 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNK 85 |||+ ||+||| | || # + |+||| || + + + Sbjct 37 KRHADGTYTSDVSTYLQ#DQAAKDFVSWLKSGRARR 71 |
| Danio rerio | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDE 94 ||||+|||++||||||+#+|||||||||||| ||| ++ ||| | Sbjct 48 KRHSEGTFSNDYSKYLE#TRRAQDFVQWLMNAKRNGGSV-KRHAE 90 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWL 78 |||++||+||| | || # + || || || Sbjct 86 KRHAEGTYTSDVSSYLQ#DQAAQSFVAWL 113 |
| Bos taurus | Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNN 87 |+++||| |||| +|# | |||| ||+ | |++ Sbjct 81 RYAEGTFISDYSIAMD#KIRQQDFVNWLLAQKGKKSD 116 |
| Macaca mulatta | Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNN 87 |+++||| |||| +|# |||| ||+ | ||+ Sbjct 51 RYAEGTFISDYSIAMD#KIHQQDFVNWLLAQKGKKND 86 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN 84 |+ ||||+| + |||#++ |+||| || + ++ Sbjct 1 HADGTFTNDMTSYLD#AKAARDFVSWLARSDKS 32 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRH 92 |++| + ||||+|||++||||||+#+|||||||||| |+|| | + +|| Sbjct 40 LTEPIEPLNMKRHSEGTFSNDYSKYLE#TRRAQDFVQWLKNSKR--NGLFRRH 89 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWL 78 +||+ ||+||| | || # + |+||| || Sbjct 87 RRHADGTYTSDVSSYLQ#DQAAKDFVSWL 114 |
| Ictalurus punctatus | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRH 92 +|||+|||++||||||+#+||||||||||||+||+ ++ |+|| Sbjct 49 RRHSEGTFSNDYSKYLE#TRRAQDFVQWLMNSKRS-DSPARRH 89 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRH 92 +|| |+|||| +| ||#| |++++||+||+ + + ||| Sbjct 134 RRHVDGSFTSDVNKVLD#SLAAKEYLQWVMNSPSSAS--GKRH 173 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKR 91 +||+ ||+||| | || # + |+||+ || + + + + | Sbjct 87 RRHADGTYTSDVSSYLQ#DQAAKDFITWLKSGQPKPDAVETR 127 |
| Heloderma horridum | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMN 80 | | ++ |||| |||||| || ++# + |++|| | Sbjct 34 DSASSESFASKIKRHSDGTFTSDLSKQME#EEAVRLFIEWLKN 75 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKR--------------- 83 +|| | |+|| |||||||||||||||||#+||||||||||||||| Sbjct 39 EPLDDSRQLNEVKRHSQGTFTSDYSKYLD#TRRAQDFVQWLMNTKRSGQQGVEEREKENLL 98 Query 84 ---NKNNIAKRHDEFER 97 + | +|+ | |+|| Sbjct 99 DQLSSNGLARHHAEYER 115 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNI 88 +||+ |||||||++ ||# |+|++||+| | + ++ Sbjct 162 RRHADGTFTSDYNQLLD#DIATQEFLKWLINQKVTQRDL 199 Query 49 EDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKN 86 | +||+ | +||| | ||+# + |++|+ ||+| + ++ Sbjct 112 EYERHADGRYTSDISSYLE#GQAAKEFIAWLVNGRGRRD 149 |
| Mus musculus | Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNN 87 |+++||| |||| +|# | |||| ||+ + |++ Sbjct 43 RYAEGTFISDYSIAMD#KIRQQDFVNWLLAQRGKKSD 78 |
| Heloderma suspectum | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKR--------------- 83 +|| | |+|| |||||||||||||||||#+||||||||||||||| Sbjct 39 EPLDDSRQLNEVKRHSQGTFTSDYSKYLD#TRRAQDFVQWLMNTKRSGQQGVEEREKENLL 98 Query 84 ---NKNNIAKRHDEFER 97 + | +|+ | |+|| Sbjct 99 DQLSSNGLARHHAEYER 115 Query 49 EDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMN 80 | +||+ | +||| | ||+# + |++|+ ||+| Sbjct 112 EYERHADGRYTSDISSYLE#GQAAKEFIAWLVN 143 |
| Protopterus dolloi | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 + + + + | |||||||| |||+| |+#++ | | || ||+||| ++|||+| || Sbjct 18 EAVDESREHTEAKRHSQGTFMSDYAKLLE#AKHALDLVQRLMSTKRT-GAVSKRHNEIER 75 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 || +||++||+||| | ||+# + +|++||| + ++ +| +| Sbjct 71 NEIERHAEGTYTSDISSYLE#GQAVNEFIRWLMKGRGRRDFSETDIEEMDR 120 Query 42 SDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 |+ | |+||+ || ||| + |+#| ++|+ ||+| | Sbjct 111 SETDIEEMDRRHADGTVTSDINSVLE#SIATREFLNWLINAK 151 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDE 94 ||||+|||++||||||+#+|||||||+||||+| | + |||| | Sbjct 48 KRHSEGTFSNDYSKYLE#TRRAQDFVEWLMNSKSNGGS-AKRHAE 90 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWL 78 |||++||+||| | +| # + ||+|| || Sbjct 86 KRHAEGTYTSDISSFLR#DQAAQNFVAWL 113 |
| Bufo marinus | Query 50 DKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNT 81 |+||+ |+||||++| ||# + ||+|+ |++|| Sbjct 107 DRRHADGSFTSDFNKALD#IKAAQEFLDWIINT 138 Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLM--NTKRNKNNIAKRHD 93 ||++|||||| + +|+# + |++|| ||+ ||| ++++ | Sbjct 62 RHAEGTFTSDMTSFLE#EKAAKEFVDWLIKGRPKRNFSDVSAADD 105 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDE 94 +||++||+|+| +++|+# + |++|+ ||+ | ++ || | Sbjct 23 ERHAEGTYTNDVTQFLE#EKAAKEFIDWLLKGIPKKQRLS-RHAE 65 |
| Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | Query 38 TDPL-SDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRH 92 |||| | ++ ||||+|||++||||| +# | ||||||||||+||+ +||| Sbjct 36 TDPLLEDLMGVSNVKRHSEGTFSNDYSKYQE#ERMAQDFVQWLMNSKRS-GAPSKRH 90 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNN 87 |||+ ||+||| | || # + |+||| || + + + + Sbjct 88 KRHADGTYTSDVSTYLQ#DQAAKDFVSWLKSGRARRES 124 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHD 93 +|| |+|||| +| ||#| |++++ |+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLD#SLAAKEYLLWVMTSKTSGKSNKRQED 177 |
| Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRH 92 || |+|||++ |||| +# | |+||+ ||||+||+ +||| Sbjct 50 KRQSEGTFSNYYSKYQE#ERMARDFLHWLMNSKRS-GAPSKRH 90 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWL 78 |||+ ||+||| | || # + |+||| || Sbjct 88 KRHADGTYTSDVSTYLQ#DQAAKDFVSWL 115 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHD 93 +|| |+|||| +| ||#| |++++ |+| +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLD#SLAAKEYLLWVMTSKTSGKSNKRQED 177 |
| Neoceratodus forsteri | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 +|+ + | |||||||| |||+| |+#+| | |||| ||||||| ++||| | || Sbjct 10 EPVDASREHTEAKRHSQGTFMSDYAKLLE#ARHALDFVQRLMNTKRN-GGVSKRHSEIER 67 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKN 86 +| +||++||+||| | ||+# + | +||+||+ + ++ Sbjct 63 SEIERHAEGTYTSDISSYLE#GQAANEFVRWLLKGRGRRD 101 Query 42 SDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 || | +||+ || |+| + |+#| ++|+ ||+|+| Sbjct 103 SDTDAEEMGRRHADGTITNDMNNVLE#SIATREFLNWLINSK 143 |
| Sebastes caurinus | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRH 92 |++| ++ ||||+|||++||||||+#+|||||||||| |+||| ++ +|| Sbjct 40 LAEPIELPNMKRHSEGTFSNDYSKYLE#TRRAQDFVQWLKNSKRN-GSLFRRH 90 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWL 78 +||+ ||+||| | || # + |++|| || Sbjct 88 RRHADGTYTSDVSSYLQ#DQAAKEFVYWL 115 |
| Tetraodon nigroviridis | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRH 92 |++| ++ ||||+|||++||||||+#+|||||||||| |+||| ++ +|| Sbjct 777 LTEPSDLSNMKRHSEGTFSNDYSKYLE#TRRAQDFVQWLKNSKRN-GSLFRRH 827 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWL 78 +||+ ||+||| | || # + |++|| || Sbjct 825 RRHADGTYTSDVSAYLQ#DQAAKEFVSWL 852 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 |+ |+|||| +| ||#+ |++|+ ||++|| Sbjct 1 HADGSFTSDINKVLD#TIAAKEFLNWLISTK 30 |
| Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN------KNNIAKRHDE 94 |||||+|||||||+||# ++|+||| ||+|||| |+ ||| + Sbjct 43 RHSQGSFTSDYSKHLD#VKQAKDFVTWLLNTKRRGVDAQAGANLEKRHSD 91 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKR 83 | +|||| |+||+| + ||# |++|++|| | Sbjct 84 NLEKRHSDGSFTNDMNVMLD#RMSAKNFLEWLKQQGR 119 |
| synthetic construct | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 |||||||||||||||||||||||||||||#|||||||||||||||||+|||||||||||| Sbjct 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNRNNIAKRHDEFER 97 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 +||+ |+|+ + + ||#+ |+||+ ||+ || Sbjct 144 RRHADGSFSDEMNTILD#NLAARDFINWLIQTK 175 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | Query 43 DPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRH 92 +| +++ ||||+|||++||||||+# |+||+||+|||| ||+ ||| Sbjct 43 EPRELSNMKRHSEGTFSNDYSKYLE#DRKAQEFVRWLMNNKRS-GVAEKRH 91 Query 50 DKRHSQGTFTSDYSKYLD#SRRAQDFVQWL 78 +|||+ |||||| | || # + +||| | Sbjct 88 EKRHADGTFTSDVSSYLK#DQAIKDFVDRL 116 |
| Sus scrofa | Query 38 TDPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 |||| ||||| |||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 38 TDPLDDPDQMTEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 +||+ |+|+ + + ||#+ +||+ ||++|| Sbjct 144 RRHADGSFSDEMNTVLD#NLATRDFINWLLHTK 175 |
| Canis lupus familiaris | Query 38 TDPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 |+||+| |||||||||||||||||||||||#|||||||||||||||||||||||||||||| Sbjct 38 TEPLNDLDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFER 97 Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 +||+ |+|+ + + ||#+ +||+ ||+ || Sbjct 144 RRHADGSFSDEMNTVLD#TLATRDFINWLLQTK 175 |
| Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRN 84 |||||||+|||||||#+|||||||||||+|||+ Sbjct 1 HSQGTFTNDYSKYLD#TRRAQDFVQWLMSTKRS 32 Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWL 78 |+ ||+||| | || # + |+ || || Sbjct 45 HADGTYTSDVSSYLQ#DQAAKKFVTWL 70 |
| Heloderma suspectum, venom, Peptide, 39 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMN 80 | +|||||| || ++# + |++|| | Sbjct 1 HGEGTFTSDLSKQME#EEAVRLFIEWLKN 28 |
| Phodopus sungorus | Query 51 KRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTK 82 +||+ |+|+ + + ||#| +||+ ||+ || Sbjct 44 RRHADGSFSDEMNTILD#SLATRDFINWLIQTK 75 Query 56 GTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKN 86 |||||| | ||+# + |++|+ ||+ + ++ Sbjct 1 GTFTSDVSSYLE#GQAAKEFIAWLVKGRGRRD 31 |
| Squalus acanthias | Query 44 PDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFE 96 |++| |+||+ |||||+ + ||#+ |++|+ |++|+| | | | | Sbjct 118 PEEM--DRRHADGTFTSEINIVLD#TIAAKEFLNWILNSK----TIQSRDSEIE 164 Query 67 D#SRRAQDFVQWLMNTKRNKNNIAKRHDE 94 |#+|||+|||||||+|||| + ||| | Sbjct 1 D#NRRAKDFVQWLMSTKRNGDK-TKRHAE 27 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWL--MNTKRNKNNIAKRHDEFER 97 ++ |||++||+||| |# +|+ || | | ++| +|+||| | | Sbjct 20 DKTKRHAEGTYTSDVDSLSD#YFKAKRFVDSLTSYNKRQNGRSISKRHVESTR 71 Query 49 EDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLM 79 | ||++|++ | | ||+#++ |+||+ ||+ Sbjct 68 ESTRHTEGSYR-DISSYLE#AKAAKDFINWLI 97 |
| Hydrolagus colliei | Query 39 DPLSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNI--AKRH 92 + +|+| + + |||+ | |+||||||||#+|| +||||||++|||| | ||| Sbjct 18 ESVSNPSTVTDVKRHTDGIFSSDYSKYLD#NRRTKDFVQWLLSTKRNGANTDKTKRH 73 Query 48 NEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNN--IAKRHDEFER 97 ++ |||+ | +||| + |# +++ ||+ | | + +|+ ++||| | | Sbjct 68 DKTKRHADGIYTSDVASLTD#YLKSKRFVESLSNYNKRQNDRRMSKRHVEGAR 119 Query 49 EDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAKRHDEFE 96 | | ++ + +|+| ||+#++ |+||+ ||+ + + + |+ | | Sbjct 116 EGARRTEEDYPNDFSSYLE#AKAAKDFIDWLIK-GQGRRDFAEESREIE 162 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | Query 56 GTFTSDYSKYLD#SRRAQDFVQWL 78 ||+||| | || # + |++|| || Sbjct 3 GTYTSDVSSYLQ#DQAAKEFVSWL 25 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | Query 53 HSQGTFTSDYSKYLD#SRRAQDFVQWLMN 80 |+ || ||| | +|+# + ++|+ ||++ Sbjct 1 HADGTLTSDISSFLE#KQATKEFIAWLVS 28 |
| Candida glabrata | Query 41 LSDPDQMNEDKRHSQGTFTSDYSKYLD#SRRAQDFVQWLMNTKRNKNNIAK 90 |+| | ||+ | | | | + +|#+ || |+ + | +| +| Sbjct 351 LADGDDDEEDEEDSSSTSESSYDEEID#NEEKQDIVEEEEDADENDDNFSK 400 |
| swine, intestines, Peptide Partial, 56 aa | Query 52 RHSQGTFTSDYSKYLD#SRRAQDFVQWLM 79 ||| |||||+ |+ |#| | | +| |+ Sbjct 29 RHSDGTFTSELSRLRD#SARLQRLLQGLV 56 |
[Site 5] RAQDFVQWLM79-NTKRNKNNIA
Met79  Asn
Asn
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Arg70 | Ala71 | Gln72 | Asp73 | Phe74 | Val75 | Gln76 | Trp77 | Leu78 | Met79 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Asn80 | Thr81 | Lys82 | Arg83 | Asn84 | Lys85 | Asn86 | Asn87 | Ile88 | Ala89 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
| DKRHSQGTFTSDYSKYLDSRRAQDFVQWLMNTKRNKNNIAKRHDEFERHAEGTFTSDVSS |
Summary
| # | organism | max score | hits | top seq |
|---|---|---|---|---|
| 1 | Homo sapiens | 122.00 | 10 | glucagon, isoform CRA_b |
| 2 | N/A | 80.90 | 45 | 0 |
| 3 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | 64.70 | 1 | AAB31092.1 |
| 4 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | 61.60 | 1 | glucagon=proglucagon-derived 3.5 kda peptide |
| 5 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | 60.80 | 1 | glucagon II |
| 6 | Contains: Glucagon; Glucagon-like peptide | 60.10 | 4 | GLUC_AMICA Glucagon precursor |
| 7 | Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | 60.10 | 1 | glucagon |
| 8 | Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | 58.50 | 1 | glucagon G-29 |
| 9 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | 57.80 | 1 | GLUC1_ORENI Glucagon-1 (Glucagon I) gb |
| 10 | Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | 56.20 | 1 | GLUC_LAMFL Glucagon gb |
| 11 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | 53.50 | 1 | glucagon |
| 12 | deshis1, Desphe6, Glu9 | 53.50 | 1 | A Chain A, Nmr Solution Structure Of The Glucagon |
| 13 | Monodelphis domestica | 50.10 | 3 | PREDICTED: similar to Gastric inhibitory polypepti |
| 14 | Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | 48.50 | 1 | GLUC1_PETMA Glucagon-1 precursor |
| 15 | Hoplobatrachus rugulosus | 46.60 | 1 | AF324209_1 proglucagon |
| 16 | Bufo marinus | 46.60 | 1 | proglucagon |
| 17 | Gallus gallus | 46.20 | 4 | glucagon isoform 1 |
| 18 | Meleagris gallopavo | 46.20 | 2 | preproglucagon B |
| 19 | Xenopus laevis | 45.80 | 5 | proglucagon II |
| 20 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | 44.70 | 1 | GLUC_HELSU Glucagon precursor |
| 21 | Rattus norvegicus | 43.90 | 6 | Glucose-dependent insulinotropic peptide |
| 22 | Canis familiaris | 43.90 | 2 | PREDICTED: similar to Gastric inhibitory polypepti |
| 23 | Ambloplites rupestris | 43.50 | 1 | proglucagon |
| 24 | Ictalurus punctatus | 43.10 | 3 | proglucagon I |
| 25 | Xenopus tropicalis | 42.70 | 2 | glucagon |
| 26 | Bos taurus | 42.40 | 5 | vasoactive intestinal peptide |
| 27 | Oncorhynchus tshawytscha | 42.40 | 1 | S78474_1 proglucagon |
| 28 | Danio rerio | 42.00 | 6 | hypothetical protein LOC494052 |
| 29 | Macaca mulatta | 42.00 | 3 | PREDICTED: gastric inhibitory polypeptide |
| 30 | Pan troglodytes | 42.00 | 2 | PREDICTED: gastric inhibitory polypeptide |
| 31 | Agkistrodon piscivorus | 41.60 | 1 | proglucagon |
| 32 | Capra hircus | 41.60 | 1 | glucagon |
| 33 | Squalus acanthias | 41.60 | 1 | proglucagon |
| 34 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | 41.20 | 1 | GLUC2_LOPAM Glucagon-2 precursor (Glucagon II) |
| 35 | Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | 41.20 | 1 | glucagon-like peptide, GLP |
| 36 | Mus musculus | 40.80 | 6 | Gastric inhibitory polypeptide |
| 37 | Sebastes caurinus | 40.80 | 4 | proglucagon I |
| 38 | Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | 40.00 | 2 | GLUC_CARAU Glucagon precursor |
| 39 | Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC2_ONCMY Glucagon-2 precursor (Glucagon II) |
| 40 | Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | 40.00 | 1 | GLUC1_ONCMY Glucagon-1 precursor (Glucagon I) |
| 41 | Heloderma horridum | 40.00 | 1 | EXE3_HELHO Exendin-3 precursor emb |
| 42 | Heloderma suspectum | 39.70 | 2 | proglucagon |
| 43 | Tetraodon nigroviridis | 39.30 | 3 | unnamed protein product |
| 44 | Protopterus dolloi | 38.90 | 1 | proglucagon |
| 45 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | 38.90 | 1 | glucagon-like peptide-2, GLP-2=proglucagon-derived |
| 46 | Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | 38.50 | 1 | VIP_MOUSE VIP peptides precursor |
| 47 | Arvicanthis ansorgei | 37.70 | 1 | vasoactive intestinal polypeptide |
| 48 | synthetic construct | 37.00 | 1 | Homo sapiens glucagon |
| 49 | Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | 37.00 | 1 | GLUC2_PETMA Glucagon-2 precursor |
| 50 | Phodopus sungorus | 36.60 | 1 | glucagon precursor |
| 51 | Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | 36.60 | 1 | GLUC1_LOPAM Glucagon-1 precursor (Glucagon I) |
| 52 | Sus scrofa | 36.20 | 1 | glucagon |
| 53 | Neoceratodus forsteri | 36.20 | 1 | proglucagon |
| 54 | Canis lupus familiaris | 35.80 | 1 | glucagon |
| 55 | Heloderma suspectum, venom, Peptide, 39 aa | 35.40 | 1 | A Chain A, Solution Structure Of Exendin-4 In 30-V |
| 56 | Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | 35.40 | 1 | GLUC_LEPSP Glucagon precursor |
| 57 | Hydrolagus colliei | 33.90 | 1 | proglucagon |
| 58 | Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | 33.50 | 1 | GLUC2_ORENI Glucagon-2 (Glucagon II) gb |
| 59 | Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | 33.50 | 1 | glucagon-like peptide-1, GLP-1=proglucagon-derived |
Top-ranked sequences
| organism | matching |
|---|---|
| Homo sapiens | Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSD 106 ||||||||||||||||||||||||||||||#|||||+||||||||||||||||||||| Sbjct 133 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSD 189 |
| N/A | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIA 89 |||||||||||||||||||||||||||#|||||||||| Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIA 89 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRN 84 ||+||||||||||+|+|||+|||||||#+|||| Sbjct 1 HSEGTFTSDYSKYMDNRRAKDFVQWLM#STKRN 32 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 |||||||||||||||+||||||+||||#+| Sbjct 1 HSQGTFTSDYSKYLDNRRAQDFIQWLM#ST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRN 84 ||+|||++||||||++|||||||||| #|+||| Sbjct 1 HSEGTFSNDYSKYLETRRAQDFVQWLK#NSKRN 32 |
| Contains: Glucagon; Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 |||||||+|||||+|+|||||||||||#+| Sbjct 1 HSQGTFTNDYSKYMDTRRAQDFVQWLM#ST 29 |
| Amia calva=bowfin, pancreatic tissues, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 |||||||+|||||+|+|||||||||||#+| Sbjct 1 HSQGTFTNDYSKYMDTRRAQDFVQWLM#ST 29 |
| Scyliorhinus canicula=European common dogfish, pancreas, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 ||+||||||||||+|+|||+|||||||#+| Sbjct 1 HSEGTFTSDYSKYMDNRRAKDFVQWLM#ST 29 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 36 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRN 84 ||+|||++||||||+ |+|||||+|||#| ||+ Sbjct 1 HSEGTFSNDYSKYLEDRKAQDFVRWLM#NNKRS 32 |
| Lampetra fluviatilis=river lamprey, small intestine, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 ||||+|||||||||||++|+||| |||#|| Sbjct 1 HSQGSFTSDYSKYLDSKQAKDFVIWLM#NT 29 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 29 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#N 80 ||+|||||||||||++++|+|||+|||#| Sbjct 1 HSEGTFTSDYSKYLENKQAKDFVRWLM#N 28 |
| deshis1, Desphe6, Glu9 | Query 54 SQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 |||| ||+||||||||||||||||||#|| Sbjct 1 SQGT-TSEYSKYLDSRRAQDFVQWLM#NT 27 |
| Monodelphis domestica | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGT 102 +|+++||| |||| +| |||| ||+#+ | || + ||+ || |+|| Sbjct 51 QRYAEGTFISDYSITMDKIMQQDFVNWLL#SQKGKKN--SWRHNITERKADGT 100 |
| Contains: Glucagon-1 (Glucagon I); Glucagon-like peptide 1-I (GLP-1I); Glucagon-like peptide 2-I (GLP-2I) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||||||||||++++|+|||+|||#| || + + + |||+||||+|++| Sbjct 41 KRHSEGTFTSDYSKYLENKQAKDFVRWLM#NAKRGGSELQR------RHADGTFTNDMTS 93 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM---#NTKRNKNNIAKRHDEFERHAE 100 +||+ ||||+| + |||++ |+||| || # ++|+ + + | +|||| Sbjct 80 RRHADGTFTNDMTSYLDAKAARDFVSWLARSD#KSRRDGGDHLAENSEDKRHAE 132 |
| Hoplobatrachus rugulosus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVS 108 |||||||||||||||||||||||||||||#|+||+ |+||+ +||||||||+|+||+ Sbjct 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NSKRS-GGISKRNVQFERHAEGTYTNDVT 107 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 +||++||+|+| +++|+ + |++|+ ||+# | | ++ ||||||||||++| Sbjct 95 ERHAEGTYTNDVTQFLEEKAAKEFIDWLI#KGKPKKQRLS-------RHAEGTFTSDMTS 146 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM--#NTKRNKNNIAKRHDEFERHAEGTFTSD 106 ||++|||||| + ||+ + |++|| ||+ # ||| +|+ + |||+|+|||| Sbjct 134 RHAEGTFTSDMTSYLEEKAAKEFVDWLIKG#RPKRNFPDISAEEMD-RRHADGSFTSD 189 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 |+||+ |+||||++| || + ||+|+ |++#|| Sbjct 178 DRRHADGSFTSDFNKALDIKAAQEFLDWII#NT 209 |
| Bufo marinus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM--#NTKRNKNNIAKRHDEFERHAEGTFTSD 106 ||++|||||| + +|+ + |++|| ||+ # ||| ++++ | |||+|+|||| Sbjct 62 RHAEGTFTSDMTSFLEEKAAKEFVDWLIKG#RPKRNFSDVSAADDMDRRHADGSFTSD 118 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 +||++||+|+| +++|+ + |++|+ ||+# | ++ ||||||||||++| Sbjct 23 ERHAEGTYTNDVTQFLEEKAAKEFIDWLL#KGIPKKQRLS-------RHAEGTFTSDMTS 74 Query 73 DFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVS 108 || ||||#|+||+ +++|+ +||||||||+|+||+ Sbjct 1 DFAQWLM#NSKRS-GGMSRRNVQFERHAEGTYTNDVT 35 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NT 81 |+||+ |+||||++| || + ||+|+ |++#|| Sbjct 107 DRRHADGSFTSDFNKALDIKAAQEFLDWII#NT 138 |
| Gallus gallus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRN------------------KNNIAKRH 92 |||||||||||||||||||||||||||||#+|||| | |+||| Sbjct 53 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#STKRNGQQGQEDKENDKFPDQLSSNAISKRH 112 Query 93 DEFERHAEGTFTSDVSS 109 |||||||||+|||++| Sbjct 113 SEFERHAEGTYTSDITS 129 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAK---RHDEFERHAEGTFTSDV 107 +||++||+||| + ||+ + |++|+ ||+#| + ++ | + |||+||||||+ Sbjct 116 ERHAEGTYTSDITSYLEGQAAKEFIAWLV#NGRGRRDFPEKALMAEEMGRRHADGTFTSDI 175 Query 108 S 108 + Sbjct 176 N 176 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNI 88 +||+ |||||| +| || |++|++||+#||| + ++ Sbjct 164 RRHADGTFTSDINKILDDMAAKEFLKWLI#NTKVTQRDL 201 |
| Meleagris gallopavo | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRN------------------KNNIAKRH 92 |||||||||||||||||||||||||||||#+|||| | |+||| Sbjct 53 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#STKRNGQQGQEDKENDEFPDQLSSNAISKRH 112 Query 93 DEFERHAEGTFTSDVSS 109 |||||||||+|||++| Sbjct 113 SEFERHAEGTYTSDITS 129 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAK---RHDEFERHAEGTFTSDV 107 +||++||+||| + ||+ + |++|+ ||+#| + ++ | + |||+||||||+ Sbjct 116 ERHAEGTYTSDITSYLEGQAAKEFIAWLV#NGRGRRDFPEKALMAEEMGRRHADGTFTSDI 175 Query 108 S 108 + Sbjct 176 N 176 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNI 88 +||+ |||||| +| || |++|++||+#||| + ++ Sbjct 164 RRHADGTFTSDINKILDDMAAKEFLKWLI#NTKVTQRDL 201 |
| Xenopus laevis | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVS 108 |||||||||||||||||||||||||||||#||||+ +++|+ ++||||||||||||+ Sbjct 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRS-GGLSRRNADYERHAEGTFTSDVT 107 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 +||++||+|+| ++||+ + + |++||+# | | + ||||||||+|+++ Sbjct 140 ERHAEGTYTNDVTEYLEEKATKAFIEWLI#KGKPKK-------IRYSRHAEGTFTNDMTN 191 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRH 92 ||++||||+| + ||+ + |++|| ||+#| + + ++ + | Sbjct 179 RHAEGTFTNDMTNYLEEKAAKEFVGWLI#NGRPKRKDLLEEH 219 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide 1 (GLP-1); Glucagon-like peptide 1(7-37) (GLP-1(7-37)); Glucagon-like peptide 1(7-36) (GLP-1(7-36)); Glucagon-like peptide 2 (GLP-2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKR------------------NKNNIAKRH 92 |||||||||||||||||+|||||||||||#|||| + | +|+ | Sbjct 51 KRHSQGTFTSDYSKYLDTRRAQDFVQWLM#NTKRSGQQGVEEREKENLLDQLSSNGLARHH 110 Query 93 DEFERHAEGTFTSDVSS 109 |+||||+| +|||+|| Sbjct 111 AEYERHADGRYTSDISS 127 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAK---RHDEFERHAEGTFTSD 106 +||+ | +||| | ||+ + |++|+ ||+#| + ++ + + | |||+|||||| Sbjct 114 ERHADGRYTSDISSYLEGQAAKEFIAWLV#NGRGRRDFLEEAGTADDIGRRHADGTFTSD 172 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNI 88 +||+ |||||||++ || |+|++||+#| | + ++ Sbjct 162 RRHADGTFTSDYNQLLDDIATQEFLKWLI#NQKVTQRDL 199 |
| Rattus norvegicus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| | |||| ||+# | ||+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDFVNWLL#AQKGKKND--WKHNLTQREA 88 |
| Canis familiaris | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| | |||| ||+# | ||+ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDFVNWLL#AQKGKKND--WKHNITQREA 88 |
| Ambloplites rupestris | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 |||||||+||+ ||+ |+||||++|||#| || + |++ |||+|||| | || Sbjct 1 HSQGTFTNDYTNYLEDRQAQDFIRWLM#NNKR--SGAAEK-----RHADGTFTDDASS 50 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK-RNKNNIAKRHDEF-ERHAEGTFTSDV 107 +|||+ |||| | | + +||| | #+ + | + +| + | || ||+||||+ Sbjct 36 EKRHADGTFTDDASSDFYDQAIKDFVAKLK#SGQDRREPETDRRREAFSRRHVEGSFTSDI 95 Query 108 S 108 + Sbjct 96 N 96 |
| Ictalurus punctatus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 +|||+|||++||||||++|||||||||||#|+||+ + |||+||+|||||| Sbjct 49 RRHSEGTFSNDYSKYLETRRAQDFVQWLM#NSKRSDS-------PARRHADGTYTSDVSS 100 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEF--ERHAEGTFTSDVS 108 +||+ ||+||| | || + |+||+ || #+ + + + | + || +|+|||||+ Sbjct 87 RRHADGTYTSDVSSYLQDQAAKDFITWLK#SGQPKPDAVETRSVDTLRRRHVDGSFTSDVN 146 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRH 92 +|| |+|||| +| ||| |++++||+|#|+ + + ||| Sbjct 134 RRHVDGSFTSDVNKVLDSLAAKEYLQWVM#NSPSSAS--GKRH 173 |
| Xenopus tropicalis | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVS 108 ||||||||||||||||||||||||+||||#||||+ +++|+ ++||||||||||||+ Sbjct 51 KRHSQGTFTSDYSKYLDSRRAQDFIQWLM#NTKRS-GGLSRRNADYERHAEGTFTSDVT 107 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 +||++||+|+| ++||+ + |++|++||+#| | | + ||||||||+|+++ Sbjct 140 ERHAEGTYTNDVTEYLEEKAAKEFIEWLI#NGKPKK-------IRYSRHAEGTFTNDMTN 191 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFE---RHAEGTFTSDVS 108 ||++||||+| + ||+ + |++|| ||+# | | | ++ | | |||+|+||+|++ Sbjct 179 RHAEGTFTNDMTNYLEEKAAKEFVGWLI#K-GRPKRNFSEAHSVEEMDRRHADGSFTNDIN 237 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNI 88 |+||+ |+||+| +| || ||+|+ |++#||+ + ++ Sbjct 224 DRRHADGSFTNDINKVLDIIAAQEFLDWVI#NTQVTERDL 262 |
| Bos taurus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSD 106 ||+ | ||||||+ | |+ +++ |+# || |+|++ +||++ || + Sbjct 80 RHADGVFTSDYSRLLGQLSAKKYLESLI#G-KRVSNSISEDQGPIKRHSDAVFTDN 133 |
| Oncorhynchus tshawytscha | Query 54 SQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 | |||++||||| + | |||||||||#|+||+ +|||+||+|||||+ Sbjct 2 SXGTFSNDYSKYQEERMAQDFVQWLM#NSKRS-------GAPSKRHADGTYTSDVST 50 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNK 85 |||+ ||+||| | || + |+||| || #+ + + Sbjct 37 KRHADGTYTSDVSTYLQDQAAKDFVSWLK#SGRARR 71 |
| Danio rerio | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++||||||++|||||||||||#| ||| ++ +||||||+|||||| Sbjct 48 KRHSEGTFSNDYSKYLETRRAQDFVQWLM#NAKRNGGSV-------KRHAEGTYTSDVSS 99 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWL 78 |||++||+||| | || + || || || Sbjct 86 KRHAEGTYTSDVSSYLQDQAAQSFVAWL 113 |
| Macaca mulatta | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| |||| ||+# | ||+ +|+ +| | Sbjct 51 RYAEGTFISDYSIAMDKIHQQDFVNWLL#AQKGKKND--WKHNITQREA 96 |
| Pan troglodytes | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| |||| ||+# | ||+ +|+ +| | Sbjct 51 RYAEGTFISDYSIAMDKIHQQDFVNWLL#AQKGKKND--WKHNITQREA 96 |
| Agkistrodon piscivorus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRH---DEFERHAEGTFTSD 106 +||+ ||+||| | ||+ + |++|+ ||+#| + ++ | | |||+|||||| Sbjct 34 ERHADGTYTSDISSYLEGQAAKEFIAWLV#NGRGRRDFSEGAHTGEDLGRRHADGTFTSD 92 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNI 88 +||+ |||||||+| || |+|++||+#| | + ++ Sbjct 82 RRHADGTFTSDYNKLLDDIATQEFLKWLI#NQKVTQRDL 119 Query 66 LDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 |+ | ++|+ | #+ | +|+ | |+||||+||+|||+|| Sbjct 9 LEDREKENFLDQLA#S-----NGLARPHAEYERHADGTYTSDISS 47 |
| Capra hircus | Query 86 NNIAKRHDEFERHAEGTFTSDVSS 109 |||||||||||||||||||||||| Query 86 NNIAKRHDEFERHAEGTFTSDVSS 109 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM# 79 +||++|||||| | ||+ + |++|+ ||+# Sbjct 11 ERHAEGTFTSDVSSYLEGQAAKEFIAWLV# 39 |
| Squalus acanthias | Query 67 DSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 |+|||+|||||||#+|||| |+ +||||||+|||| | Sbjct 1 DNRRAKDFVQWLM#STKRN-------GDKTKRHAEGTYTSDVDS 36 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWL--M#NTKRNKNNIAKRHDEFERHAEGTFTSDVS 108 |||++||+||| | +|+ || | #| ++| +|+||| | || ||++ |+| Sbjct 23 KRHAEGTYTSDVDSLSDYFKAKRFVDSLTSY#NKRQNGRSISKRHVESTRHTEGSY-RDIS 81 Query 109 S 109 | Query 109 S 109 Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKN---------NIAKRHDEFERHAEGT 102 ||++|++ | | ||+++ |+||+ ||+# + + | + |||+|| Sbjct 71 RHTEGSYR-DISSYLEAKAAKDFINWLI#KGRGRREFPEESKEIVNEVIPEEMDRRHADGT 129 Query 103 FTSDVS 108 |||+++ Sbjct 130 FTSEIN 135 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFE 96 |+||+ |||||+ + ||+ |++|+ |++#|+| | | | | Sbjct 122 DRRHADGTFTSEINIVLDTIAAKEFLNWIL#NSK----TIQSRDSEIE 164 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-2 (Glucagon II); Glucagon-like peptide 2 (Glucagon-like peptide II) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++||||||++|||||||||| #|+||| | |||+||+|||||| Sbjct 50 KRHSEGTFSNDYSKYLETRRAQDFVQWLK#NSKRN--------GLFRRHADGTYTSDVSS 100 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWL 78 +||+ ||+||| | || + |+||| || Sbjct 87 RRHADGTYTSDVSSYLQDQAAKDFVSWL 114 |
| Petromyzon marinus=sea lampreys, intestine, parasitic phase, Peptide, 32 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRN 84 |+ ||||+| + |||++ |+||| || # + ++ Sbjct 1 HADGTFTNDMTSYLDAKAARDFVSWLA#RSDKS 32 |
| Mus musculus | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHA 99 |+++||| |||| +| | |||| ||+# + |++ +|+ +| | Sbjct 43 RYAEGTFISDYSIAMDKIRQQDFVNWLL#AQRGKKSD--WKHNITQREA 88 |
| Sebastes caurinus | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++|||+||+ |+|||||+|||#| ||+ || +|||+||||||||| Sbjct 48 KRHSEGTFSNDYSRYLEERKAQDFVRWLM#NNKRS------GADE-KRHADGTFTSDVSS 99 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK-RNKNNIAKRHDEF-ERHAEGTFTSDV 107 +|||+ |||||| | || + +||| | #+ + | ++ | + | || +|+||||| Sbjct 85 EKRHADGTFTSDVSSYLKDQAIKDFVNRLK#SGQVRRESETEWRGEAFSRRHVDGSFTSDV 144 Query 108 S 108 + Sbjct 145 N 145 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDE 94 +|| |+|||| +| ||| |++++ |+|# +| + + ++ |+ Sbjct 133 RRHVDGSFTSDVNKVLDSMAAKEYLLWVM#TSKPSGESKKRQEDQ 176 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon; Glucagon-like peptide | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++||||||++|||||||+|||#|+| | + +||||||+|||+|| Sbjct 48 KRHSEGTFSNDYSKYLETRRAQDFVEWLM#NSKSNGGSA-------KRHAEGTYTSDISS 99 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWL 78 |||++||+||| | +| + ||+|| || Sbjct 86 KRHAEGTYTSDISSFLRDQAAQNFVAWL 113 |
| Contains: Glicentin-related polypeptide 2 (GRPP 2); Glucagon-2; Glucagon-like peptide 1-2 (GLP 1-2); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 || |+|||++ |||| + | |+||+ |||#|+||+ +|||+||+|||||+ Sbjct 50 KRQSEGTFSNYYSKYQEERMARDFLHWLM#NSKRS-------GAPSKRHADGTYTSDVST 101 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#N--TKRNKNNIAKRHDEFERHAEGTFTSDVS 108 |||+ ||+||| | || + |+||| || #+ +| + || +|+|||||+ Sbjct 88 KRHADGTYTSDVSTYLQDQAAKDFVSWLK#SGPARRESAEESMNGPMSRRHVDGSFTSDVN 147 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHD 93 +|| |+|||| +| ||| |++++ |+|# +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSLAAKEYLLWVM#TSKTSGKSNKRQED 177 |
| Contains: Glicentin-related polypeptide 1 (GRPP 1); Glucagon-1; Glucagon-like peptide 1-1 (GLP 1-1); Glucagon-like peptide 2 (GLP 2) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++||||| + | |||||||||#|+||+ +|||+||+|||||+ Sbjct 50 KRHSEGTFSNDYSKYQEERMAQDFVQWLM#NSKRS-------GAPSKRHADGTYTSDVST 101 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#N--TKRNKNNIAKRHDEFERHAEGTFTSDVS 108 |||+ ||+||| | || + |+||| || #+ +| ++ || +|+|||||+ Sbjct 88 KRHADGTYTSDVSTYLQDQAAKDFVSWLK#SGRARRESAEESRNGPMSRRHVDGSFTSDVN 147 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHD 93 +|| |+|||| +| ||| |++++ |+|# +| + + ++ | Sbjct 135 RRHVDGSFTSDVNKVLDSLAAKEYLLWVM#TSKTSGKSNKRQED 177 |
| Heloderma horridum | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#N 80 |||| |||||| || ++ + |++|| #| Sbjct 46 KRHSDGTFTSDLSKQMEEEAVRLFIEWLK#N 75 |
| Heloderma suspectum | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKR------------------NKNNIAKRH 92 |||||||||||||||||+|||||||||||#|||| + | +|+ | Sbjct 51 KRHSQGTFTSDYSKYLDTRRAQDFVQWLM#NTKRSGQQGVEEREKENLLDQLSSNGLARHH 110 Query 93 DEFERHAEGTFTSDVSS 109 |+||||+| +|||+|| Sbjct 111 AEYERHADGRYTSDISS 127 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#N 80 +||+ | +||| | ||+ + |++|+ ||+#| Sbjct 114 ERHADGRYTSDISSYLEGQAAKEFIAWLV#N 143 |
| Tetraodon nigroviridis | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++||||||++|||||||||| #|+||| + | |||+||+|||||+ Sbjct 787 KRHSEGTFSNDYSKYLETRRAQDFVQWLK#NSKRNGS-------LFRRHADGTYTSDVSA 838 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWL 78 +||+ ||+||| | || + |++|| || Sbjct 825 RRHADGTYTSDVSAYLQDQAAKEFVSWL 852 |
| Protopterus dolloi | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 |||||||| |||+| |+++ | | || ||#+||| ++|||+| |||||||+|||+|| Sbjct 30 KRHSQGTFMSDYAKLLEAKHALDLVQRLM#STKRT-GAVSKRHNEIERHAEGTYTSDISS 87 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFE-RHAEGTFTSDVSS 109 +||++||+||| | ||+ + +|++|||# + ++ +| + |||+|| |||++| Sbjct 74 ERHAEGTYTSDISSYLEGQAVNEFIRWLM#KGRGRRDFSETDIEEMDRRHADGTVTSDINS 133 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 |+||+ || ||| + |+| ++|+ ||+#| | Sbjct 119 DRRHADGTVTSDINSVLESIATREFLNWLI#NAK 151 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 33 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 |+ |+|||| +| ||+ |++|+ ||+#+|| Sbjct 1 HADGSFTSDINKVLDTIAAKEFLNWLI#STK 30 |
| Contains: Intestinal peptide PHI-42; Intestinal peptide PHI-27 (Peptide histidine isoleucinamide 27); Vasoactive intestinal peptide (VIP) (Vasoactive intestinal polypeptide) | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSD 106 ||+ | ||||||+ | |+ +++ |+# || ++|++ +||++ || + Sbjct 80 RHADGVFTSDYSRLLGQISAKKYLESLI#G-KRISSSISEDPVPIKRHSDAVFTDN 133 |
| Arvicanthis ansorgei | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSD 106 ||+ | ||||||+ | |+ +++ |+# || ++| + +||++ || + Sbjct 63 RHADGVFTSDYSRLLGQISAKKYLESLI#G-KRISSSILEDPVPIKRHSDAVFTDN 116 |
| synthetic construct | Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 ||||||||||||||||||||||||||||||#|||||+|||||||||||||||||||||||| Sbjct 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNRNNIAKRHDEFERHAEGTFTSDVSS 109 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKN---NIAKRHDEFERHAEGTFTSDV 107 +||++|||||| | ||+ + |++|+ ||+# + ++ +| + |||+|+|+ ++ Sbjct 96 ERHAEGTFTSDVSSYLEGQAAKEFIAWLV#KGRGRRDFPEEVAIVEELGRRHADGSFSDEM 155 Query 108 SS 109 ++ Sbjct 156 NT 157 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 +||+ |+|+ + + ||+ |+||+ ||+# || Sbjct 144 RRHADGSFSDEMNTILDNLAARDFINWLI#QTK 175 |
| Contains: Glucagon-2 (Glucagon II) (Glu II); Glucagon-like peptide 2-II (GLP-2II) | Query 52 RHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVS 108 |||||+|||||||+|| ++|+||| ||+#|||| + + +||++|+||+|++ Sbjct 43 RHSQGSFTSDYSKHLDVKQAKDFVTWLL#NTKRRGVDAQAGANLEKRHSDGSFTNDMN 99 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKR 83 +|||| |+||+| + || |++|++|| # | Sbjct 86 EKRHSDGSFTNDMNVMLDRMSAKNFLEWLK#QQGR 119 |
| Phodopus sungorus | Query 56 GTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKN---NIAKRHDEFERHAEGTFTSDVSS 109 |||||| | ||+ + |++|+ ||+# + ++ + + |||+|+|+ ++++ Sbjct 1 GTFTSDVSSYLEGQAAKEFIAWLV#KGRGRRDFPEEVTIVEELGRRHADGSFSDEMNT 57 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 +||+ |+|+ + + ||| +||+ ||+# || Sbjct 44 RRHADGSFSDEMNTILDSLATRDFINWLI#QTK 75 |
| Contains: Glicentin-related polypeptide (GRPP); Glucagon-1 (Glucagon I); Glucagon-like peptide 1 (Glucagon-like peptide I) | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 ||||+|||++||||||+ |+||+||+|||#| || + +|++ |||+||||||||| Sbjct 51 KRHSEGTFSNDYSKYLEDRKAQEFVRWLM#NNKR--SGVAEK-----RHADGTFTSDVSS 102 Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWL 78 +|||+ |||||| | || + +||| | Sbjct 88 EKRHADGTFTSDVSSYLKDQAIKDFVDRL 116 |
| Sus scrofa | Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKN---NIAKRHDEFERHAEGTFTSDV 107 +||++|||||| | ||+ + |++|+ ||+# + ++ + + |||+|+|+ ++ Sbjct 96 ERHAEGTFTSDVSSYLEGQAAKEFIAWLV#KGRGRRDFPEEVTIVEELRRRHADGSFSDEM 155 Query 108 SS 109 ++ Sbjct 156 NT 157 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 +||+ |+|+ + + ||+ +||+ ||+#+|| Sbjct 144 RRHADGSFSDEMNTVLDNLATRDFINWLL#HTK 175 |
| Neoceratodus forsteri | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 |||||||| |||+| |++| | |||| ||#||||| ++||| | |||||||+|||+|| Sbjct 22 KRHSQGTFMSDYAKLLEARHALDFVQRLM#NTKRN-GGVSKRHSEIERHAEGTYTSDISS 79 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEF-ERHAEGTFTSDVSS 109 +||++||+||| | ||+ + | +||+||+# + ++ +| |||+|| |+|+++ Sbjct 66 ERHAEGTYTSDISSYLEGQAANEFVRWLL#KGRGRRDFSDTDAEEMGRRHADGTITNDMNN 125 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 +||+ || |+| + |+| ++|+ ||+#|+| Sbjct 112 RRHADGTITNDMNNVLESIATREFLNWLI#NSK 143 |
| Canis lupus familiaris | Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 ||||||||||||||||||||||||||||||#|||||||||||||||||||||||||||||| Query 50 DKRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKN---NIAKRHDEFERHAEGTFTSDV 107 +||++|||||| | ||+ + |++|+ ||+# + ++ +| + |||+|+|+ ++ Sbjct 96 ERHAEGTFTSDVSSYLEGQAAKEFIAWLV#KGRGRRDFPEEVAIVEEFRRRHADGSFSDEM 155 Query 108 SS 109 ++ Sbjct 156 NT 157 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTK 82 +||+ |+|+ + + ||+ +||+ ||+# || Sbjct 144 RRHADGSFSDEMNTVLDTLATRDFINWLL#QTK 175 |
| Heloderma suspectum, venom, Peptide, 39 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#N 80 | +|||||| || ++ + |++|| #| Sbjct 1 HGEGTFTSDLSKQMEEEAVRLFIEWLK#N 28 |
| Contains: Glucagon; Glucagon-36 (Oxyntomodulin); Glucagon-like peptide | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 |||||||+|||||||+|||||||||||#+|||+ | ||+||+|||||| Sbjct 1 HSQGTFTNDYSKYLDTRRAQDFVQWLM#STKRS-GGITXXXXXXXXHADGTYTSDVSS 56 Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWL 78 |+ ||+||| | || + |+ || || Sbjct 45 HADGTYTSDVSSYLQDQAAKKFVTWL 70 |
| Hydrolagus colliei | Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFERHAEGTFTSDVSS 109 |||+ | |+||||||||+|| +||||||+#+|||| | |+ +|||+| +||||+| Sbjct 30 KRHTDGIFSSDYSKYLDNRRTKDFVQWLL#STKRNGANT----DKTKRHADGIYTSDVAS 84 Query 51 KRHSQGTFTSDYSKYLDSRRAQDFVQWLM#NTKRNKNN--IAKRHDEFERHAEGTFTSDVS 108 |||+ | +||| + | +++ ||+ | #| + +|+ ++||| | | | + +| | Sbjct 71 KRHADGIYTSDVASLTDYLKSKRFVESLS#NYNKRQNDRRMSKRHVEGARRTEEDYPNDFS 130 Query 109 S 109 | Query 109 S 109 Query 51 KRHSQGT------FTSDYSKYLDSRRAQDFVQWLM#NTKRNKNNIAKRHDEFE 96 ||| +| + +|+| ||+++ |+||+ ||+# + + + |+ | | Sbjct 112 KRHVEGARRTEEDYPNDFSSYLEAKAAKDFIDWLI#K-GQGRRDFAEESREIE 162 |
| Tilapia nilotica=Nile tilapia, Brockmann body, Peptide, 33 aa | Query 56 GTFTSDYSKYLDSRRAQDFVQWL 78 ||+||| | || + |++|| || Sbjct 3 GTYTSDVSSYLQDQAAKEFVSWL 25 |
| Amphiuma tridactylum=three-toed amphiumae, pancreas, Peptide, 34 aa | Query 53 HSQGTFTSDYSKYLDSRRAQDFVQWLM#N 80 |+ || ||| | +|+ + ++|+ ||+#+ Sbjct 1 HADGTLTSDISSFLEKQATKEFIAWLV#S 28 |
* References
None.