XSB2251 : PREDICTED: similar to Gap junction alpha-8 protein (Connexin-49) (Cx49) (Lens fiber protein MP70) (MP64) (MP38) [Equus caballus]
[ CaMP Format ]
This entry is computationally expanded from SB0016
* Basic Information
| Organism | Equus caballus (horse) |
| Protein Names | similar to Gap junction alpha-8 protein (Connexin-49) (Cx49) (Lens fiber protein MP70) (MP64) (MP38) |
| Gene Names | LOC100065084; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 21 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
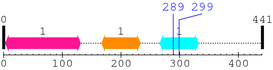
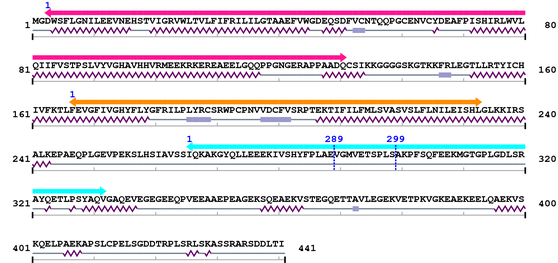
Length: 441 aa
Average Mass: 48.959 kDa
Monoisotopic Mass: 48.928 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
| Connexin 1. | 3 | 131 | 311.7 | 1.5e-90 |
| Connexin_CCC 1. | 167 | 233 | 166.7 | 6.6e-47 |
| Connexin50 1. | 266 | 332 | 181.2 | 2.9e-51 |
| --- cleavage 289 (inside Connexin50 266..332) --- |
| --- cleavage 299 (inside Connexin50 266..332) --- |
3. Sequence Information
Fasta Sequence: XSB2251.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
2 [sites]
Cleavage sites (±10aa)
[Site 1] IVSHYFPLAE289-VGMVETSPLS
Glu289  Val
Val
 |
| P10 | P9 | P8 | P7 | P6 |
P5 | P4 | P3 | P2 | P1 |
| Ile280 | Val281 | Ser282 | His283 | Tyr284 | Phe285 | Pro286 | Leu287 | Ala288 | Glu289 |
 |
| P1' | P2' | P3' | P4' | P5' |
P6' | P7' | P8' | P9' | P10' |
| Val290 | Gly291 | Met292 | Val293 | Glu294 | Thr295 | Ser296 | Pro297 | Leu298 | Ser299 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
|
SIAVSSIQKAKGYQLLEEEKIVSHYFPLAEVGMVETSPLSAKPFSQFEEKMGTGPLGDLS
|
Summary
| # |
organism |
max score |
hits |
top seq |
| 1 |
Canis familiaris |
118.00 |
6 |
PREDICTED: similar to gap junction membrane channe |
| 2 |
Cricetulus griseus |
113.00 |
1 |
connexin50 |
| 3 |
Rattus norvegicus |
113.00 |
1 |
gap junction membrane channel protein alpha 8 |
| 4 |
Mus musculus |
113.00 |
1 |
gap junction membrane channel protein alpha 8 |
| 5 |
Mesocricetus auratus |
113.00 |
1 |
connexin50 |
| 6 |
N/A |
112.00 |
1 |
C |
| 7 |
Ovis aries |
112.00 |
1 |
AF177913_1 connexin 49 protein |
| 8 |
Bos taurus |
112.00 |
1 |
PREDICTED: similar to Gap junction alpha-8 protein |
| 9 |
Homo sapiens |
111.00 |
3 |
gap junction protein, alpha 8, 50kDa (connexin 50) |
| 10 |
Pan troglodytes |
111.00 |
1 |
PREDICTED: gap junction protein, alpha 8, 50kDa (c |
| 11 |
Monodelphis domestica |
105.00 |
1 |
PREDICTED: similar to Gap junction alpha-8 protein |
| 12 |
Macaca mulatta |
94.70 |
1 |
PREDICTED: gap junction protein, alpha 8, 50kDa (c |
| 13 |
Gallus gallus |
79.70 |
1 |
gap junction protein, alpha 8, 50kDa (connexin 50) |
| 14 |
Danio rerio |
60.80 |
3 |
AF288817_1 gap junction protein connexin 44.1 |
| 15 |
Tetraodon nigroviridis |
49.30 |
1 |
unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|
| Canis familiaris |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 319
|||||||||||||||||||||||||||| |#||||||||||||||||||||||||||||||
Sbjct 261 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 320
|
| Cricetulus griseus |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 319
|||||||||||||||||||||||||||| |#||||||||||||||||||||+||||| |+|
Sbjct 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
|
| Rattus norvegicus |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 319
|||||||||||||||||||||||||||| |#||||||||||||||||||||+||||| |+|
Sbjct 261 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 320
|
| Mus musculus |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 319
|||||||||||||||||||||||||||| |#||||||||||||||||||||+||||| |+|
Sbjct 261 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 320
|
| Mesocricetus auratus |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 319
|||||||||||||||||||||||||||| |#||||||||||||||||||||+||||| |+|
Sbjct 243 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLSAKPFSQFEEKIGTGPLADMS 302
|
| N/A |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 319
|||||||||||||||||||||||||||| |#||||| ||||||||||||||+| |||||||
Sbjct 261 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVEASPLSAKPFSQFEEKVGPGPLGDLS 320
|
| Ovis aries |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 319
|||||||||||||||||||||||||||| |#||||| ||||||||||||||+| |||||||
Sbjct 261 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVEASPLSAKPFSQFEEKVGPGPLGDLS 320
|
| Bos taurus |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 319
|||||||||||||||||||||||||||| |#||||| ||||||||||||||+| |||||||
Sbjct 261 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVEASPLSAKPFSQFEEKVGPGPLGDLS 320
|
| Homo sapiens |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 319
|||||||||||||||||||||||||||| |#||||||||| ||||+|||||+ ||||||||
Sbjct 255 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLPAKPFNQFEEKISTGPLGDLS 314
|
| Pan troglodytes |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 319
|||||||||||||||||||||||||||| |#||||||||| ||||+|||||+ ||||||||
Sbjct 254 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVETSPLPAKPFNQFEEKISTGPLGDLS 313
|
| Monodelphis domestica |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 319
|||||||||||||||||||||||||||| |#||+||| ||+| |||+||||+ ||||||||
Sbjct 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGVVETRPLAATPFSRFEEKISTGPLGDLS 319
|
| Macaca mulatta |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 319
|||||||||||||||||||||||||||| |#|||||| ||||||||||+ ||| |||
Sbjct 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLTE#VGMVET----AKPFSQFEEKITTGPPEDLS 315
|
| Gallus gallus |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEKMGTGPLGDLS 319
+|||||| |||||+|||||| ||||||| |#|| || ||| + |++||||+| ||| |||
Sbjct 256 AIAVSSIPKAKGYKLLEEEKPVSHYFPLTE#VG-VEPSPLPS-AFNEFEEKIGMGPLEDLS 313
|
| Danio rerio |
Query 260 SIAVSSIQKAKGYQLLEEEKIVSHYFPLAE#VGMVETSPLSAKPFSQFEEK 309
||| |||||||||+||||+| ||+||| |#|| +| | | + ||||
Sbjct 262 SIAASSIQKAKGYKLLEEDKSTSHFFPLTE#VGGMEAGRLPAS-YEPFEEK 310
|
| Tetraodon nigroviridis |
Query 260 SIAVSSIQKAKGYQLLEEEKI--VSHYFPLAE#VGMVETSPLSAKPFSQFEEK 309
|+|| |+|+ |||+|||||| ++| +||||#||| + + || |||
Sbjct 268 SLAVPSMQRVKGYRLLEEEKAPPITHLYPLAE#VGM--EAGRGSPPFQGLEEK 317
|
[Site 2] VGMVETSPLS299-AKPFSQFEEK
Ser299  Ala
Ala
 |
| P10 | P9 | P8 | P7 | P6 |
P5 | P4 | P3 | P2 | P1 |
| Val290 | Gly291 | Met292 | Val293 | Glu294 | Thr295 | Ser296 | Pro297 | Leu298 | Ser299 |
 |
| P1' | P2' | P3' | P4' | P5' |
P6' | P7' | P8' | P9' | P10' |
| Ala300 | Lys301 | Pro302 | Phe303 | Ser304 | Gln305 | Phe306 | Glu307 | Glu308 | Lys309 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
|
KGYQLLEEEKIVSHYFPLAEVGMVETSPLSAKPFSQFEEKMGTGPLGDLSRAYQETLPSY
|
Summary
| # |
organism |
max score |
hits |
top seq |
| 1 |
Canis familiaris |
122.00 |
6 |
PREDICTED: similar to gap junction membrane channe |
| 2 |
Rattus norvegicus |
116.00 |
1 |
gap junction membrane channel protein alpha 8 |
| 3 |
Cricetulus griseus |
116.00 |
1 |
connexin50 |
| 4 |
N/A |
116.00 |
1 |
C |
| 5 |
Mus musculus |
116.00 |
1 |
gap junction membrane channel protein alpha 8 |
| 6 |
Ovis aries |
116.00 |
1 |
AF177913_1 connexin 49 protein |
| 7 |
Bos taurus |
116.00 |
1 |
PREDICTED: similar to Gap junction alpha-8 protein |
| 8 |
Homo sapiens |
114.00 |
3 |
gap junction protein, alpha 8, 50kDa (connexin 50) |
| 9 |
Mesocricetus auratus |
114.00 |
1 |
connexin50 |
| 10 |
Pan troglodytes |
114.00 |
1 |
PREDICTED: gap junction protein, alpha 8, 50kDa (c |
| 11 |
Monodelphis domestica |
107.00 |
1 |
PREDICTED: similar to Gap junction alpha-8 protein |
| 12 |
Macaca mulatta |
97.40 |
1 |
PREDICTED: gap junction protein, alpha 8, 50kDa (c |
| 13 |
Gallus gallus |
81.60 |
1 |
gap junction protein, alpha 8, 50kDa (connexin 50) |
| 14 |
Danio rerio |
61.60 |
3 |
AF288817_1 gap junction protein connexin 44.1 |
| 15 |
Tetraodon nigroviridis |
55.10 |
1 |
unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|
| Canis familiaris |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 329
|||||||||||||||||| |||||||||||#||||||||||||||||||||||||||||||
Sbjct 271 KGYQLLEEEKIVSHYFPLTEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 330
|
| Rattus norvegicus |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 329
|||||||||||||||||| |||||||||||#||||||||||+||||| |+|| ||||||||
Sbjct 271 KGYQLLEEEKIVSHYFPLTEVGMVETSPLS#AKPFSQFEEKIGTGPLADMSRGYQETLPSY 330
|
| Cricetulus griseus |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 329
|||||||||||||||||| |||||||||||#||||||||||+||||| |+|| ||||||||
Sbjct 253 KGYQLLEEEKIVSHYFPLTEVGMVETSPLS#AKPFSQFEEKIGTGPLADMSRGYQETLPSY 312
|
| N/A |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 329
|||||||||||||||||| |||||| ||||#||||||||||+| |||||||||||||||||
Sbjct 271 KGYQLLEEEKIVSHYFPLTEVGMVEASPLS#AKPFSQFEEKVGPGPLGDLSRAYQETLPSY 330
|
| Mus musculus |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 329
|||||||||||||||||| |||||||||||#||||||||||+||||| |+||+||||||||
Sbjct 271 KGYQLLEEEKIVSHYFPLTEVGMVETSPLS#AKPFSQFEEKIGTGPLADMSRSYQETLPSY 330
|
| Ovis aries |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 329
|||||||||||||||||| |||||| ||||#||||||||||+| |||||||||||||||||
Sbjct 271 KGYQLLEEEKIVSHYFPLTEVGMVEASPLS#AKPFSQFEEKVGPGPLGDLSRAYQETLPSY 330
|
| Bos taurus |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 329
|||||||||||||||||| |||||| ||||#||||||||||+| |||||||||||||||||
Sbjct 271 KGYQLLEEEKIVSHYFPLTEVGMVEASPLS#AKPFSQFEEKVGPGPLGDLSRAYQETLPSY 330
|
| Homo sapiens |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 329
|||||||||||||||||| |||||||||| #||||+|||||+ ||||||||| ||||||||
Sbjct 265 KGYQLLEEEKIVSHYFPLTEVGMVETSPLP#AKPFNQFEEKISTGPLGDLSRGYQETLPSY 324
|
| Mesocricetus auratus |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 329
|||||||||||||||||| |||||||||||#||||||||||+||||| |+|| | ||||||
Sbjct 253 KGYQLLEEEKIVSHYFPLTEVGMVETSPLS#AKPFSQFEEKIGTGPLADMSRGYPETLPSY 312
|
| Pan troglodytes |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 329
|||||||||||||||||| |||||||||| #||||+|||||+ ||||||||| ||||||||
Sbjct 264 KGYQLLEEEKIVSHYFPLTEVGMVETSPLP#AKPFNQFEEKISTGPLGDLSRGYQETLPSY 323
|
| Monodelphis domestica |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 329
|||||||||||||||||| |||+||| ||+#| |||+||||+ ||||||||||| ||||||
Sbjct 270 KGYQLLEEEKIVSHYFPLTEVGVVETRPLA#ATPFSRFEEKISTGPLGDLSRAYTETLPSY 329
|
| Macaca mulatta |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 329
|||||||||||||||||| ||||||| #||||||||||+ ||| |||| ||||||||
Sbjct 270 KGYQLLEEEKIVSHYFPLTEVGMVET----#AKPFSQFEEKITTGPPEDLSRGYQETLPSY 325
|
| Gallus gallus |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTGPLGDLSRAYQETLPSY 329
|||+|||||| ||||||| ||| || ||| #+ |++||||+| ||| |||||+ | ||||
Sbjct 266 KGYKLLEEEKPVSHYFPLTEVG-VEPSPLP#SA-FNEFEEKIGMGPLEDLSRAFDERLPSY 323
|
| Danio rerio |
Query 270 KGYQLLEEEKIVSHYFPLAEVGMVETSPLS#AKPFSQFEEKM--GTGPLGDLSRAYQETLP 327
|||+||||+| ||+||| ||| +| | #| + |||| | |+|+ | ||||
Sbjct 272 KGYKLLEEDKSTSHFFPLTEVGGMEAGRLP#AS-YEPFEEKSDEAMAPKKDMSKMYDETLP 330
Query 328 SY 329
||
Query 328 SY 329
|
| Tetraodon nigroviridis |
Query 270 KGYQLLEEEKI--VSHYFPLAEVGMVETSPLS#AKPFSQFEEKMGTG-PLGDLSRAYQETL 326
|||+|||||| ++| +||||||| + #+ || ||| |+ |+|+ | |||
Sbjct 278 KGYRLLEEEKAPPITHLYPLAEVGM--EAGRG#SPPFQGLEEKPEEVLPMEDISKVYDETL 335
Query 327 PSY 329
|||
Query 327 PSY 329
|
 Val
Val

 Sequence conservation (by blast)
Sequence conservation (by blast) Ala
Ala

 Sequence conservation (by blast)
Sequence conservation (by blast)