XSB2803 : PREDICTED: similar to beta-B3 crystallin [Sus scrofa]
[ CaMP Format ]
This entry is computationally expanded from SB0068
* Basic Information
| Organism | Sus scrofa (pig) |
| Protein Names | similar to beta-B3 crystallin |
| Gene Names | LOC100157434; Derived by automated computational analysis using gene prediction method: GNOMON. Supporting evidence includes similarity to: 32 ESTs, 5 Proteins |
| Gene Locus | Not available |
| GO Function | Not available |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
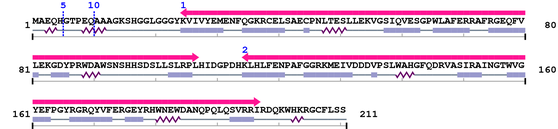
Length: 211 aa
Average Mass: 24.256 kDa
Monoisotopic Mass: 24.241 kDa
2. Domain Information
Annotated Domains: Not Available.
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
| --- cleavage 10 --- |
| --- cleavage 5 --- |
| Crystall 1. | 25 | 107 | 159.9 | 7.7e-45 |
| Crystall 2. | 115 | 197 | 160.5 | 5.2e-45 |
3. Sequence Information
Fasta Sequence: XSB2803.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
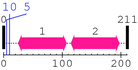
2 [sites]
Cleavage sites (±10aa)
[Site 1] MAEQHGTPEQ10-AAAGKSHGGL
Gln10  Ala
Ala
 |
| P10 | P9 | P8 | P7 | P6 |
P5 | P4 | P3 | P2 | P1 |
| Met1 | Ala2 | Glu3 | Gln4 | His5 | Gly6 | Thr7 | Pro8 | Glu9 | Gln10 |
 |
| P1' | P2' | P3' | P4' | P5' |
P6' | P7' | P8' | P9' | P10' |
| Ala11 | Ala12 | Ala13 | Gly14 | Lys15 | Ser16 | His17 | Gly18 | Gly19 | Leu20 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
|
MAEQHGTPEQAAAGKSHGGLGGGYKVIVYEMENFQGKRCE
|
Summary
| # |
organism |
max score |
hits |
top seq |
| 1 |
Pan troglodytes |
55.50 |
2 |
PREDICTED: crystallin, beta B3 isoform 1 |
| 2 |
Bos taurus |
54.30 |
1 |
crystallin, beta B3 |
| 3 |
Homo sapiens |
52.40 |
4 |
CRBB3_HUMAN Beta crystallin B3 (Beta-B3-crystallin |
| 4 |
Ovis aries |
52.40 |
1 |
crystallin, beta B3 |
| 5 |
Macaca mulatta |
51.60 |
2 |
PREDICTED: crystallin, beta B2 isoform 1 |
| 6 |
Rattus norvegicus |
51.60 |
2 |
AF287304_1 betaB3-crystallin |
| 7 |
N/A |
51.60 |
1 |
C |
| 8 |
Mus musculus |
51.60 |
1 |
crystallin, beta B3 |
| 9 |
Cavia porcellus |
51.60 |
1 |
betaB3-crystallin |
| 10 |
Gallus gallus |
42.40 |
1 |
crystallin, beta B3 |
| 11 |
Xenopus laevis |
42.00 |
2 |
MGC84287 protein |
| 12 |
Danio rerio |
36.20 |
1 |
beta B3-crystallin |
| 13 |
Tetraodon nigroviridis |
33.50 |
1 |
unnamed protein product |
Top-ranked sequences
| organism | matching |
|---|
| Pan troglodytes |
Query 1 MAEQHGTPEQ#XXXXXXXXXXXXXYKVIVYEMENFQGKRCE 40
||||||||||# |||||||+|||||||||
Sbjct 1 MAEQHGTPEQ#AAAGKSHGGLGGSYKVIVYELENFQGKRCE 40
|
| Bos taurus |
Query 1 MAEQHGTPEQ#XXXXXXXXXXXXXYKVIVYEMENFQGKRCE 40
||||| ||||# |||||||||||||||||
Sbjct 1 MAEQHSTPEQ#AAAGKSHGGLGGSYKVIVYEMENFQGKRCE 40
|
| Homo sapiens |
Query 1 MAEQHGTPEQ#XXXXXXXXXXXXXYKVIVYEMENFQGKRCE 40
|||||| |||# ||||+||+|||||||||
Sbjct 1 MAEQHGAPEQ#AAAGKSHGDLGGSYKVILYELENFQGKRCE 40
|
| Ovis aries |
Query 1 MAEQHGTPEQ#XXXXXXXXXXXXXYKVIVYEMENFQGKRCE 40
||||| |||# |||||||||||||||||
Sbjct 1 MAEQHSAPEQ#AAAGKSHGGLGGSYKVIVYEMENFQGKRCE 40
|
| Macaca mulatta |
Query 1 MAEQHGTPEQ#XXXXXXXXXXXXXYKVIVYEMENFQGKRCE 40
|||||| |||# |||||||+|||||| ||
Sbjct 1 MAEQHGAPEQ#AAAGKSHGGLGGSYKVIVYELENFQGKHCE 40
|
| Rattus norvegicus |
Query 1 MAEQHGTPEQ#XXXXXXXXXXXXXYKVIVYEMENFQGKRCE 40
|||||| |||# ||| |||+|||||||||
Sbjct 1 MAEQHGAPEQ#AAASKSHGGLGGSYKVTVYELENFQGKRCE 40
|
| N/A |
Query 1 MAEQHGTPEQ#XXXXXXXXXXXXXYKVIVYEMENFQGKRCE 40
|||||| |||# ||| |||+|||||||||
Sbjct 1 MAEQHGAPEQ#AAASKSHGGLGGSYKVTVYELENFQGKRCE 40
|
| Mus musculus |
Query 1 MAEQHGTPEQ#XXXXXXXXXXXXXYKVIVYEMENFQGKRCE 40
|||||| |||# ||| |||+|||||||||
Sbjct 1 MAEQHGAPEQ#AAAGKSHGGLGGSYKVTVYELENFQGKRCE 40
|
| Cavia porcellus |
Query 1 MAEQHGTPEQ#XXXXXXXXXXXXXYKVIVYEMENFQGKRCE 40
|||||| |||# ||| |||+|||||||||
Sbjct 1 MAEQHGAPEQ#AAAGKSHGGLGGGYKVTVYELENFQGKRCE 40
|
| Gallus gallus |
Query 1 MAEQHGTPEQ#XXXXXXXXXXXXXYKVIVYEMENFQGKRCE 40
| || |||# ||+ +||+|||||+|||
Sbjct 1 MTEQQSPPEQ#MVTGEGAGERGGNYKITIYELENFQGRRCE 40
|
| Xenopus laevis |
Query 1 MAEQHGTPEQ#XXXXXXXXXXXXXYKVIVYEMENFQGKRCE 40
|+| ||||# ||+ +||+||||||+||
Sbjct 1 MSEPQSTPEQ#MASGKSHGGIGGNYKITLYELENFQGKKCE 40
|
| Danio rerio |
Query 1 MAEQHGTPEQ#XXXXXXXXXXXXXYKVIVYEMENFQGKRCE 40
|++| | |+|# ||| +|| |||+||+ |
Sbjct 1 MSDQQGVPDQ#QVAGKSQGGAGAIYKVSLYEFENFRGKKLE 40
|
| Tetraodon nigroviridis |
Query 1 MAEQHGTPEQ#XXXXXXXXXXXXXYKVIVYEMENFQGKRCE 40
|+|+ |||# |||+++| ||||| + |
Sbjct 33 MSERQSAPEQ#LAAGKSQGGAGATYKVLLFEFENFQGCKAE 72
|
[Site 2] MAEQH5-GTPEQAAAGK
His5  Gly
Gly
 |
| P10 | P9 | P8 | P7 | P6 |
P5 | P4 | P3 | P2 | P1 |
| - | - | - | - | - | Met1 | Ala2 | Glu3 | Gln4 | His5 |
 |
| P1' | P2' | P3' | P4' | P5' |
P6' | P7' | P8' | P9' | P10' |
| Gly6 | Thr7 | Pro8 | Glu9 | Gln10 | Ala11 | Ala12 | Ala13 | Gly14 | Lys15 |
 Sequence conservation (by blast)
Sequence conservation (by blast) Sequence conservation (by blast)
Sequence conservation (by blast)
| Reference peptide (cleaved bond±30 residues) |
|---|
|
MAEQHGTPEQAAAGKSHGGLGGGYKVIVYEMENFQ
|
Summary
| # |
organism |
max score |
hits |
top seq |
| 1 |
Pan troglodytes |
43.90 |
2 |
PREDICTED: crystallin, beta B3 isoform 1 |
| 2 |
Bos taurus |
42.70 |
1 |
crystallin, beta B3 |
| 3 |
Macaca mulatta |
42.00 |
2 |
PREDICTED: crystallin, beta B2 isoform 1 |
| 4 |
Homo sapiens |
40.80 |
4 |
CRBB3_HUMAN Beta crystallin B3 (Beta-B3-crystallin |
| 5 |
Ovis aries |
40.80 |
1 |
crystallin, beta B3 |
| 6 |
N/A |
40.00 |
1 |
C |
| 7 |
Mus musculus |
40.00 |
1 |
crystallin, beta B3 |
| 8 |
Cavia porcellus |
40.00 |
1 |
betaB3-crystallin |
| 9 |
Rattus norvegicus |
40.00 |
1 |
AF287304_1 betaB3-crystallin |
Top-ranked sequences
| organism | matching |
|---|
| Pan troglodytes |
Query 1 MAEQH#GTPEQXXXXXXXXXXXXXYKVIVYEMENFQ 35
|||||#||||| |||||||+||||
Sbjct 1 MAEQH#GTPEQAAAGKSHGGLGGSYKVIVYELENFQ 35
|
| Bos taurus |
Query 1 MAEQH#GTPEQXXXXXXXXXXXXXYKVIVYEMENFQ 35
|||||# |||| ||||||||||||
Sbjct 1 MAEQH#STPEQAAAGKSHGGLGGSYKVIVYEMENFQ 35
|
| Macaca mulatta |
Query 1 MAEQH#GTPEQXXXXXXXXXXXXXYKVIVYEMENFQ 35
|||||#| ||| |||||||+||||
Sbjct 1 MAEQH#GAPEQAAAGKSHGGLGGSYKVIVYELENFQ 35
|
| Homo sapiens |
Query 1 MAEQH#GTPEQXXXXXXXXXXXXXYKVIVYEMENFQ 35
|||||#| ||| ||||+||+||||
Sbjct 1 MAEQH#GAPEQAAAGKSHGDLGGSYKVILYELENFQ 35
|
| Ovis aries |
Query 1 MAEQH#GTPEQXXXXXXXXXXXXXYKVIVYEMENFQ 35
|||||# ||| ||||||||||||
Sbjct 1 MAEQH#SAPEQAAAGKSHGGLGGSYKVIVYEMENFQ 35
|
| N/A |
Query 1 MAEQH#GTPEQXXXXXXXXXXXXXYKVIVYEMENFQ 35
|||||#| ||| ||| |||+||||
Sbjct 1 MAEQH#GAPEQAAASKSHGGLGGSYKVTVYELENFQ 35
|
| Mus musculus |
Query 1 MAEQH#GTPEQXXXXXXXXXXXXXYKVIVYEMENFQ 35
|||||#| ||| ||| |||+||||
Sbjct 1 MAEQH#GAPEQAAAGKSHGGLGGSYKVTVYELENFQ 35
|
| Cavia porcellus |
Query 1 MAEQH#GTPEQXXXXXXXXXXXXXYKVIVYEMENFQ 35
|||||#| ||| ||| |||+||||
Sbjct 1 MAEQH#GAPEQAAAGKSHGGLGGGYKVTVYELENFQ 35
|
| Rattus norvegicus |
Query 1 MAEQH#GTPEQXXXXXXXXXXXXXYKVIVYEMENFQ 35
|||||#| ||| ||| |||+||||
Sbjct 1 MAEQH#GAPEQAAASKSHGGLGGSYKVTVYELENFQ 35
|
 Ala
Ala

 Sequence conservation (by blast)
Sequence conservation (by blast) Gly
Gly

 Sequence conservation (by blast)
Sequence conservation (by blast)