| # |
organism |
max score |
hits |
top seq |
| 1 |
Homo sapiens |
126.00 |
38 |
calpain, small subunit 1 |
| 2 |
Mus musculus |
126.00 |
33 |
calpain small subunit |
| 3 |
N/A |
126.00 |
27 |
S |
| 4 |
Rattus norvegicus |
126.00 |
13 |
calpain, small subunit 1 |
| 5 |
Bos taurus |
126.00 |
8 |
calpain, small subunit 1 |
| 6 |
Sus scrofa |
126.00 |
7 |
calpain, small subunit 1 |
| 7 |
Ovis aries |
125.00 |
3 |
calpain II small subunit |
| 8 |
Oryctolagus cuniculus |
125.00 |
2 |
calpain, small subunit 1 |
| 9 |
Gallus gallus |
123.00 |
8 |
calpain small subunit |
| 10 |
Canis familiaris |
110.00 |
23 |
PREDICTED: similar to calpain small subunit 2 |
| 11 |
Pan troglodytes |
110.00 |
8 |
PREDICTED: acyltransferase like 1 |
| 12 |
Macaca mulatta |
110.00 |
4 |
PREDICTED: similar to calpain small subunit 2 |
| 13 |
Xenopus laevis |
109.00 |
13 |
LOC446963 protein |
| 14 |
Xenopus tropicalis |
108.00 |
6 |
calpain, small subunit 2 |
| 15 |
Danio rerio |
105.00 |
26 |
hypothetical protein LOC550598 |
| 16 |
Monodelphis domestica |
103.00 |
4 |
PREDICTED: similar to Calpain, small subunit 2 |
| 17 |
Oncorhynchus mykiss |
98.20 |
5 |
calpain regulatory subunit |
| 18 |
Tetraodon nigroviridis |
94.00 |
10 |
unnamed protein product |
| 19 |
Coturnix coturnix |
80.10 |
1 |
quail calpain |
| 20 |
Macaca fascicularis |
79.30 |
1 |
CAN3_MACFA Calpain-3 (Calpain L3) (Calpain p94) (C |
| 21 |
synthetic construct |
77.40 |
1 |
Homo sapiens calpain 1, (mu/I) large subunit |
| 22 |
Pongo pygmaeus |
76.30 |
1 |
CAN1_PONPY Calpain-1 catalytic subunit (Calpain-1 |
| 23 |
Stizostedion vitreum vitreum |
74.30 |
1 |
putative calpain-like protein |
| 24 |
catalytic |
70.10 |
1 |
Calpain 2, large |
| 25 |
Strongylocentrotus purpuratus |
65.10 |
1 |
PREDICTED: similar to calpain B isoform 2 |
| 26 |
Schistosoma japonicum |
59.70 |
2 |
unknown |
| 27 |
Tribolium castaneum |
57.80 |
1 |
PREDICTED: similar to CG8107-PA |
| 28 |
Gecarcinus lateralis |
57.00 |
1 |
calpain B |
| organism | matching |
|---|
| Homo sapiens |
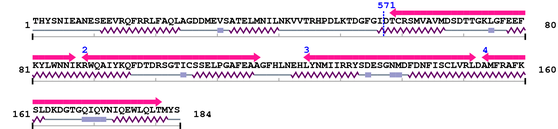
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|
| Mus musculus |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|
| N/A |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|
| Rattus norvegicus |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|
| Bos taurus |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|
| Sus scrofa |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
||||||||||||||||||||||||||||||#||||||||||||||||||||||||||||||
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|
| Ovis aries |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|||||||||||||||||||||||||||||+#||||||||||||||||||||||||||||||
Sbjct 28 DMEVSATELMNILNKVVTRHPDLKTDGFGV#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|
| Oryctolagus cuniculus |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|||||||||||||||||||||||||||||+#||||||||||||||||||||||||||||||
Sbjct 110 DMEVSATELMNILNKVVTRHPDLKTDGFGL#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 169
|
| Gallus gallus |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
||||| |||||||||||||||||||||||+#||||||||||||||||||||||||||||||
Sbjct 58 DMEVSPTELMNILNKVVTRHPDLKTDGFGL#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 117
|
| Canis familiaris |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|||| ||+||||||||+++| ||||||| +#|||||+|+||||||||||||||||||||||
Sbjct 95 DMEVGATDLMNILNKVLSKHKDLKTDGFSL#DTCRSIVSVMDSDTTGKLGFEEFKYLWNNI 154
|
| Pan troglodytes |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|||| ||+||||||||+++| ||||||| +#|||||+|+||||||||||||||||||||||
Sbjct 492 DMEVGATDLMNILNKVLSKHKDLKTDGFSL#DTCRSIVSVMDSDTTGKLGFEEFKYLWNNI 551
|
| Macaca mulatta |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|||| ||+||||||||+++| ||||||| +#|||||+|+||||||||||||||||||||||
Sbjct 92 DMEVGATDLMNILNKVLSKHKDLKTDGFSL#DTCRSIVSVMDSDTTGKLGFEEFKYLWNNI 151
|
| Xenopus laevis |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|||||| ||| |||||| || ||||||| +#|+|||||||||||+|||||||||||||+||
Sbjct 71 DMEVSAAELMGILNKVVARHQDLKTDGFSV#DSCRSMVAVMDSDSTGKLGFEEFKYLWDNI 130
|
| Xenopus tropicalis |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|||||||||| |||||+ +| ||||||| #|+|||||||||||+|||||||||||||+||
Sbjct 78 DMEVSATELMGILNKVIAKHQDLKTDGFSA#DSCRSMVAVMDSDSTGKLGFEEFKYLWDNI 137
|
| Danio rerio |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
||||| |||||||||++++| ||||||| |#++|||||||||||+|||||| |||+|||||
Sbjct 60 DMEVSPTELMNILNKIISKHGDLKTDGFTI#ESCRSMVAVMDSDSTGKLGFHEFKHLWNNI 119
|
| Monodelphis domestica |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|||| +||||||||+++| +| |||| +#|||||+||||||| ||||||||||||||||
Sbjct 89 DMEVGPADLMNILNKVISKHRELNTDGFSL#DTCRSIVAVMDSDATGKLGFEEFKYLWNNI 148
|
| Oncorhynchus mykiss |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
||||| ||||||||+++ + ||||||| |#++|| ||||||||+|||||| | |+|||||
Sbjct 60 DMEVSPTELMNILNRIIGKRSDLKTDGFSI#ESCRGMVAVMDSDSTGKLGFHELKFLWNNI 119
|
| Tetraodon nigroviridis |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
||||| || +|||||+ | ++||||| |#++|||||||||||+|||||| |||||| ||
Sbjct 61 DMEVSPGELKDILNKVIGRFSNMKTDGFSI#ESCRSMVAVMDSDSTGKLGFHEFKYLWGNI 120
|
| Coturnix coturnix |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|||+| || |||+|+ || ||||||| +#|+||+|| +|| | + +|| ||+ ||| |
Sbjct 549 DMEISVFELKTILNRVIARHKDLKTDGFSL#DSCRNMVNLMDKDGSARLGLVEFQILWNKI 608
|
| Macaca fascicularis |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|||+ | || +|| || +| |||| || +#++||||+|+||+| +||| +|| +||| |
Sbjct 659 DMEICADELKKVLNTVVNKHKDLKTHGFTL#ESCRSMIALMDTDGSGKLNLQEFHHLWNKI 718
|
| synthetic construct |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|||+| || |||+++++| ||+| || +#++||||| +|| | |||| || ||| |
Sbjct 558 DMEISVKELRTILNRIISKHKDLRTKGFSL#ESCRSMVNLMDRDGNGKLGLVEFNILWNRI 617
|
| Pongo pygmaeus |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
||| | || |||+++++| ||+| || +#++||||| +|| | |||| || ||| |
Sbjct 558 DMETSVKELRTILNRIISKHKDLRTKGFSL#ESCRSMVNLMDRDGNGKLGLVEFNILWNRI 617
|
| Stizostedion vitreum vitreum |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
| |+|+ || |||+|||+ |+|| || |# |||+|| ++| | +|||| ||| || |
Sbjct 546 DCEISSYELQKILNRVVTKRSDIKTSGFSI#TTCRNMVNMLDKDGSGKLGLLEFKVLWTKI 605
|
| catalytic |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
| |+|| || || +|+ + |+|+||| |#+||+ || ++||| +|||| +|| || |
Sbjct 574 DAEISAFELQTILRRVLAKRQDIKSDGFSI#ETCKIMVDMLDSDGSGKLGLKEFYILWTKI 633
|
| Strongylocentrotus purpuratus |
Query 28 DMEVSATELMNILNKVVTRH----PDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYL 83
|||| | || ||| + + +|+ || +#++|+||||+ | | +||||||||+ |
Sbjct 529 DMEVDAWELQEILNAALRKDLQVPGELQGGGFSL#ESCKSMVALTDDDRSGKLGFEEFREL 588
Query 84 WNNI 87
| ||
Sbjct 589 WQNI 592
|
| Schistosoma japonicum |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWN 85
| |+ | || +||| || | ||| +#++||||+|+|| | +| | | ||+ ||+
Sbjct 201 DGEIDADELRDILNCAFTR--DFTFDGFSL#ESCRSMIAMMDFDRSGMLSFPEFRKLWD 256
|
| Tribolium castaneum |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
| || || ||+ || + || #| ||||+|++| | |||| ||||| || +|
Sbjct 960 DEEVDWKELKEILDHY-TRKVKTQNRGFSK#DVCRSMIAMLDVDRTGKLNFEEFKRLWESI 1018
|
| Gecarcinus lateralis |
Query 28 DMEVSATELMNILNKVVTRHPDLKTDGFGI#DTCRSMVAVMDSDTTGKLGFEEFKYLWNNI 87
|+|+ || ++|| + | + +|| #| ||||+|+|| | +|||| +|| || +|
Sbjct 600 DLEIDWKELQDVLNFALKR--EFNFEGFSK#DVCRSMIAMMDVDRSGKLGLQEFLQLWMDI 657
|
 Asp
Asp

 Sequence conservation (by blast)
Sequence conservation (by blast)