XSB2913 : PTP 35 protein [Mus musculus]
[ CaMP Format ]
This entry is computationally expanded from SB0024
* Basic Information
| Organism | Mus musculus (house mouse) |
| Protein Names | Receptor-type tyrosine-protein phosphatase-like N; R-PTP-N; PTP IA-2; PTP 35 protein |
| Gene Names | Ptp35 |
| Gene Locus | Not available |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| CAA52453 | N/A | N/A | PTPRN_MOUSE | N/A | N/A | N/A | mmu: |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 996 aa
Average Mass: 107.659 kDa
Monoisotopic Mass: 107.592 kDa
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Toxin_19 1. | 57 | 89 | -6.3 | 2.9 |
| TrmB 1. | 365 | 421 | -30.4 | 4.9 |
| FDX-ACB 1. | 491 | 567 | -53.8 | 2.9 |
| FCD 1. | 600 | 722 | -29.6 | 8.3 |
| --- cleavage 675 (inside FCD 600..722) --- | ||||
| Y_phosphatase 1. | 751 | 985 | 386.2 | 5.8e-113 |
3. Sequence Information
Fasta Sequence: XSB2913.fasta
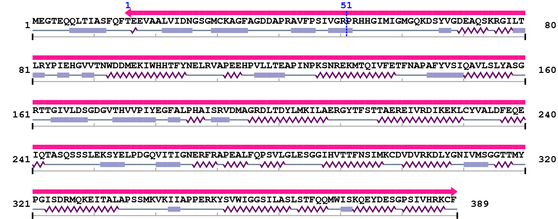
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] PEPSRVSSVS675-SQFSDAAQAS
Ser675  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Pro666 | Glu667 | Pro668 | Ser669 | Arg670 | Val671 | Ser672 | Ser673 | Val674 | Ser675 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser676 | Gln677 | Phe678 | Ser679 | Asp680 | Ala681 | Ala682 | Gln683 | Ala684 | Ser685 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 8526904] Magistrelli G, Covini N, Mosca M, Lippoli G, Isacchi A, Expression of PTP35, the murine homologue of the protein tyrosine phosphatase-related sequence IA-2, is regulated during cell growth and stimulated by mitogens in 3T3 fibroblasts. Biochem Biophys Res Commun. 1995 Dec 5;217(1):154-61.