XSB2977 : phospholipase C, beta 1 isoform 2 [Mus musculus]
[ CaMP Format ]
This entry is computationally expanded from SB0008
* Basic Information
| Organism | Mus musculus (house mouse) |
| Protein Names | Phospholipase C, beta 1; Phospholipase C, beta 1, isoform CRA_a; phospholipase C; beta 1 isoform 2; phospholipase C-beta-1a; phosphoinositide phospholipase C |
| Gene Names | Plcb1; RP23-418D21.3-003, mCG_16884; phospholipase C, beta 1 |
| Gene Locus | 2 76.7 cM; chromosome 2 |
| GO Function | Not available |
| Entrez Protein | Entrez Nucleotide | Entrez Gene | UniProt | OMIM | HGNC | HPRD | KEGG |
|---|---|---|---|---|---|---|---|
| NP_062651 | NM_019677 | 18795 | A2AW58_MOUSE | N/A | N/A | N/A | mmu:18795 |
* Information From OMIM
Not Available.
* Structure Information
1. Primary Information
Length: 1173 aa
Average Mass: 133.327 kDa
Monoisotopic Mass: 133.243 kDa
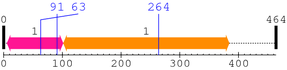
2. Domain Information
Annotated Domains: interpro / pfam / smart / prosite
Computationally Assigned Domains (Pfam+HMMER):
| domain name | begin | end | score | e-value |
|---|---|---|---|---|
| Chitin_synth_1 1. | 92 | 181 | -112.3 | 0.67 |
| efhand_like 1. | 224 | 315 | 164.0 | 4.4e-46 |
| PI-PLC-X 1. | 318 | 468 | 327.6 | 2.6e-95 |
| PI-PLC-Y 1. | 539 | 656 | 297.6 | 2.6e-86 |
| C2 1. | 678 | 761 | 59.0 | 1.7e-14 |
| --- cleavage 880 --- | ||||
| DUF1154 1. | 903 | 946 | 6.1 | 0.0037 |
| PLC-beta_C 1. | 997 | 1166 | 310.5 | 3.4e-90 |
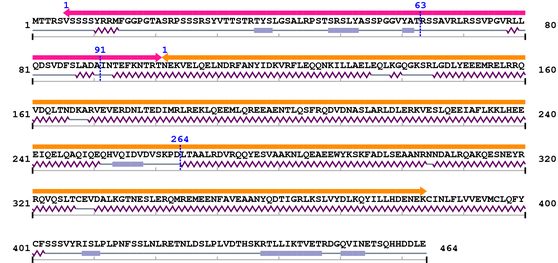
3. Sequence Information
Fasta Sequence: XSB2977.fasta
Amino Acid Sequence and Secondary Structures (PsiPred):
4. 3D Information
Not Available.
* Cleavage Information
1 [sites]
Cleavage sites (±10aa)
[Site 1] APKPPSQAPH880-SQPAPGSVKA
His880  Ser
Ser
 |
|||||||||
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 |
|---|---|---|---|---|---|---|---|---|---|
| Ala871 | Pro872 | Lys873 | Pro874 | Pro875 | Ser876 | Gln877 | Ala878 | Pro879 | His880 |
 |
|||||||||
| P1' | P2' | P3' | P4' | P5' | P6' | P7' | P8' | P9' | P10' |
| Ser881 | Gln882 | Pro883 | Ala884 | Pro885 | Gly886 | Ser887 | Val888 | Lys889 | Ala890 |
 Sequence conservation (by blast)
Sequence conservation (by blast)
* References
[PubMed ID: 19385066] O'Carroll SJ, Mitchell MD, Faenza I, Cocco L, Gilmour RS, Nuclear PLCbeta1 is required for 3T3-L1 adipocyte differentiation and regulates expression of the cyclin D3-cdk4 complex. Cell Signal. 2009 Jun;21(6):926-35.